Data export plugins¶
Isolate record export¶
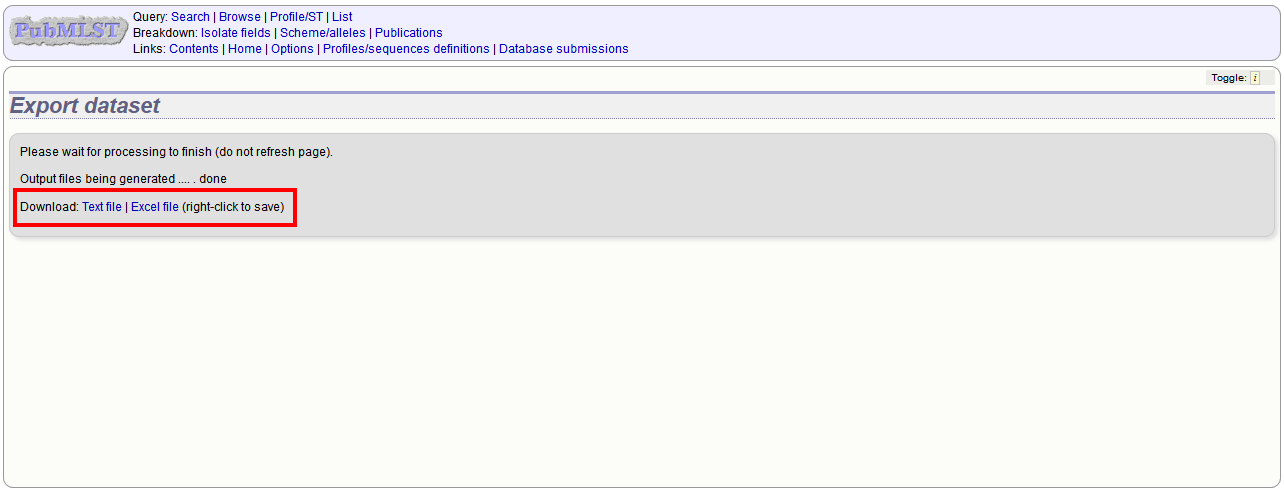
You can export the entire isolate recordset by clicking the ‘Export dataset’ link in the Export section of the main contents page.
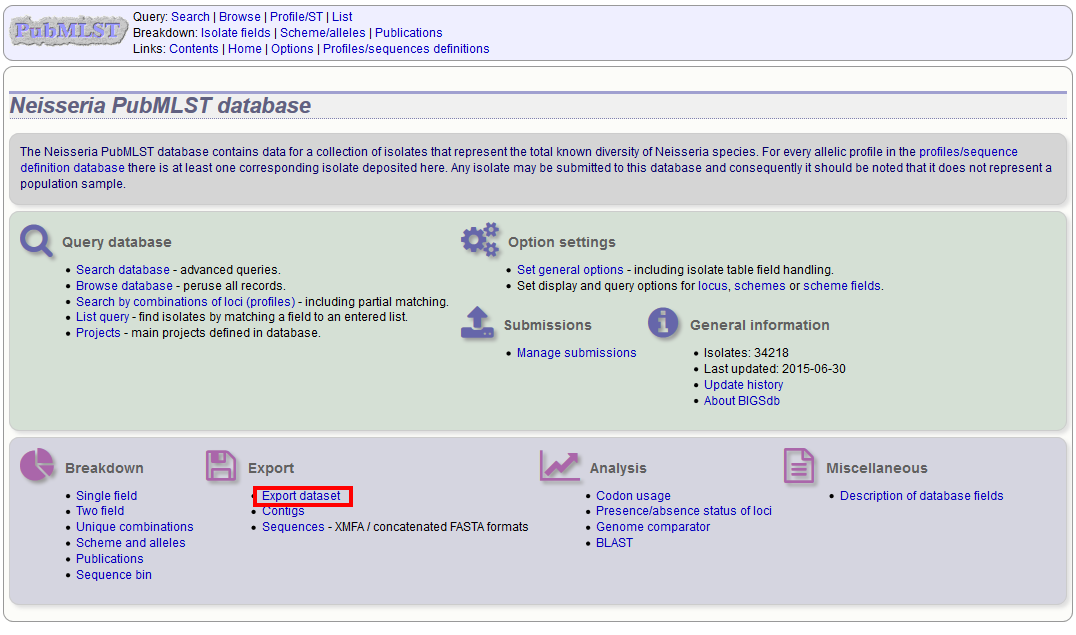
Alternatively, you can export the recordsets of isolates returned from a database query by clicking the ‘Dataset’ button in the Export list at the bottom of the results table. Please note that the list of functions here may vary depending on the setup of the database.
Select the isolate fields and schemes to include.
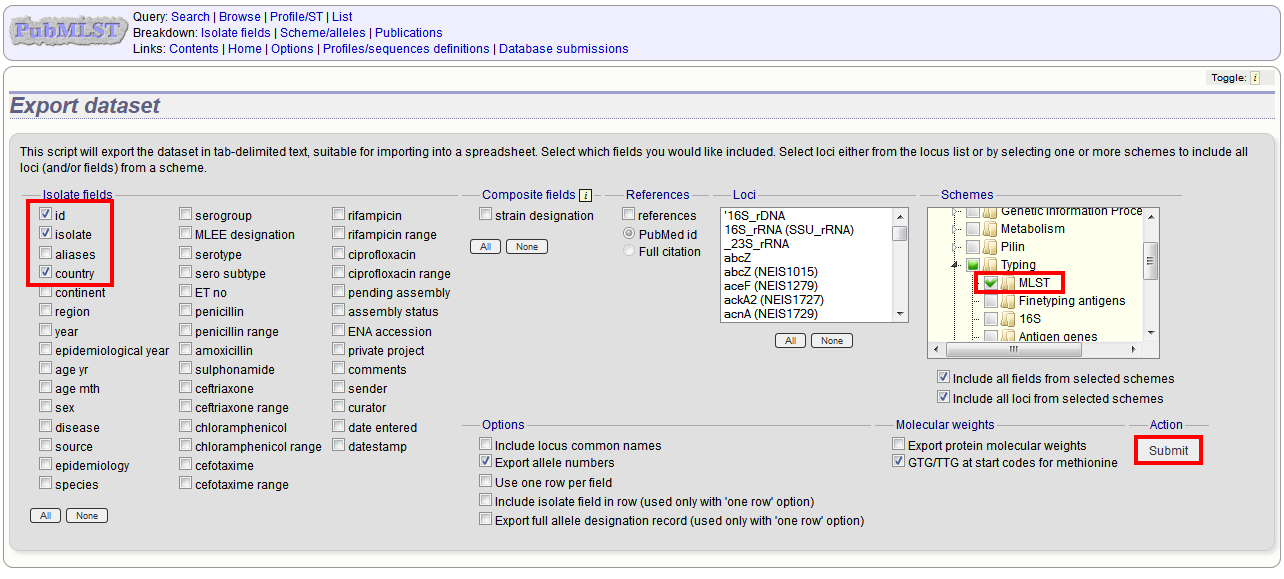
Click Submit.
You can then download the data in tab-delimited text or Excel formats.
Advanced options¶
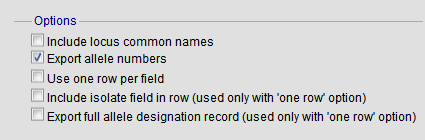
The options fieldset has the following options.
- Include locus common names - any common name for the locus is displayed in parentheses following the primary name.
- Export allele numbers - the allele designation is included for any locus included.
- Use one row per field - this is an alternative output format where instead of each locus and field having a separate column, each field is export on a separate row.
- Include isolate field in row - the name of the isolate is included as a separate column when exporting in ‘one row per field’ fomrmat.
- Export full allele designation record - export sender, curator and datestamp information as separate rows when exporting allele designation data.
Molecular weight calculation¶
The plugin can also calculate the predicted molecular weight of the gene product of any allele designated in the dataset.

Click the ‘Export protein molecular weight’ checkbox. Additional columns (or rows depending on the output format) will be created to include the molecular weight data.
Sequence export¶
You can export the sequences for any set of loci designated in isolate records, or belonging to scheme profiles in the sequence definition database.
The sequence export function can be accessed by clicking the ‘Sequences’ link in the Export section of the contents page.
Alternatively, you can access this function by clicking the ‘Sequences’ button in the Export list at the bottom of the results table. Please note that the list of functions here may vary depending on the setup of the database.
Select the isolate or profile records to analyse - these will be pre-selected if you accessed the plugin following a query. Select the loci to include either directly within the loci list and/or using the schemes tree.
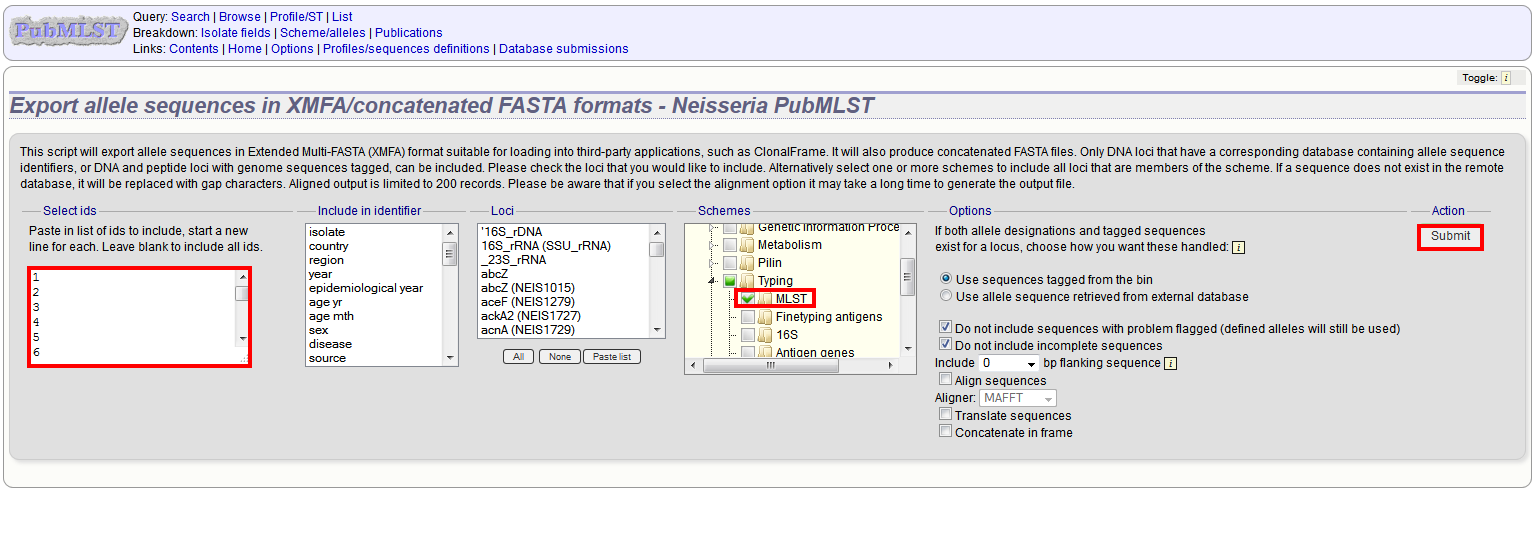
Click submit.
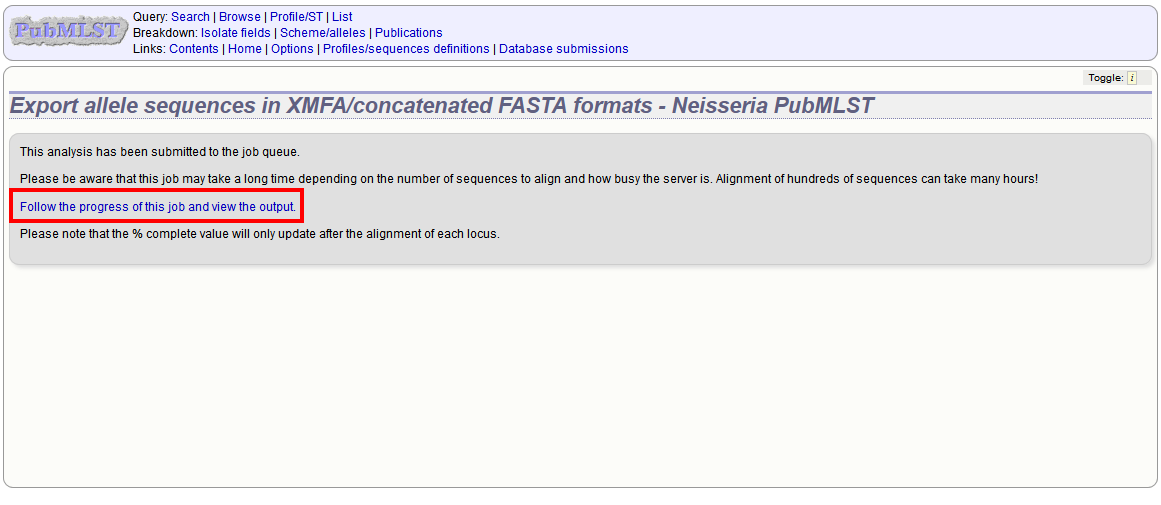
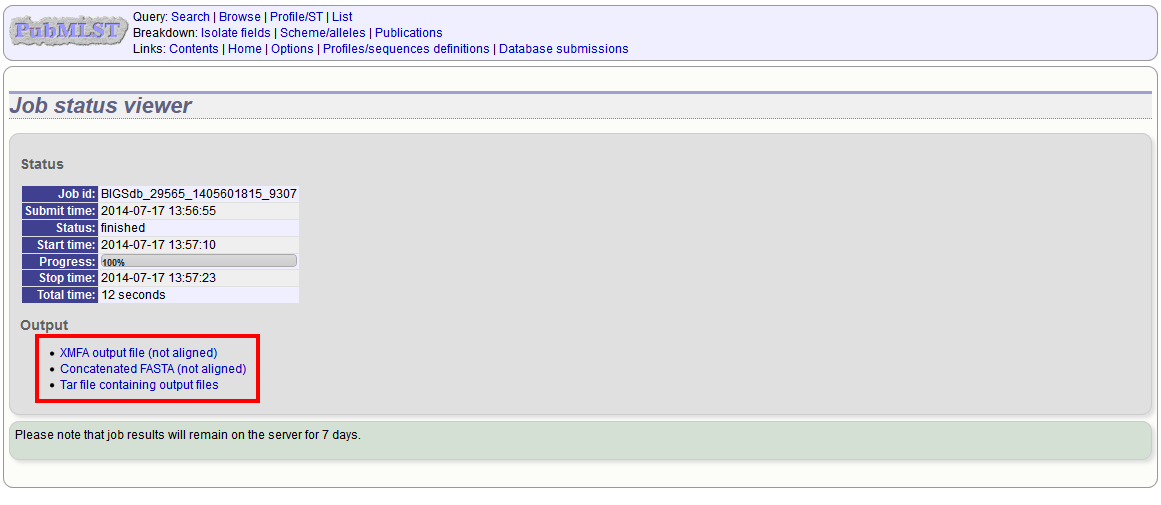
The job will be submitted to the job queue. Click the link to follow the progress and download the resulting files.
Sequences will be export in XMFA and FASTA file formats.
Aligning sequences¶
By default, sequences will be exported unaligned - this is very quick since no processing is required. You can choose to align the sequences by checking the ‘Align sequences’ checkbox.
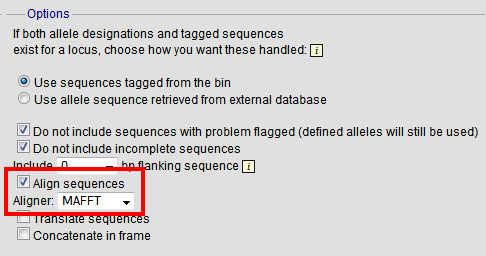
You can also choose to use MUSCLE or MAFFT as the aligner. MAFFT is the default choice and is usually much quicker than MUSCLE. Both produce comparable results.
Profile export¶
You can export the allelic profiles for any indexed scheme (those containing a primary key field) defined in the sequence definition database.
The profile export function can be accessed by clicking the ‘Profiles’ link in the Export section of the contents page.
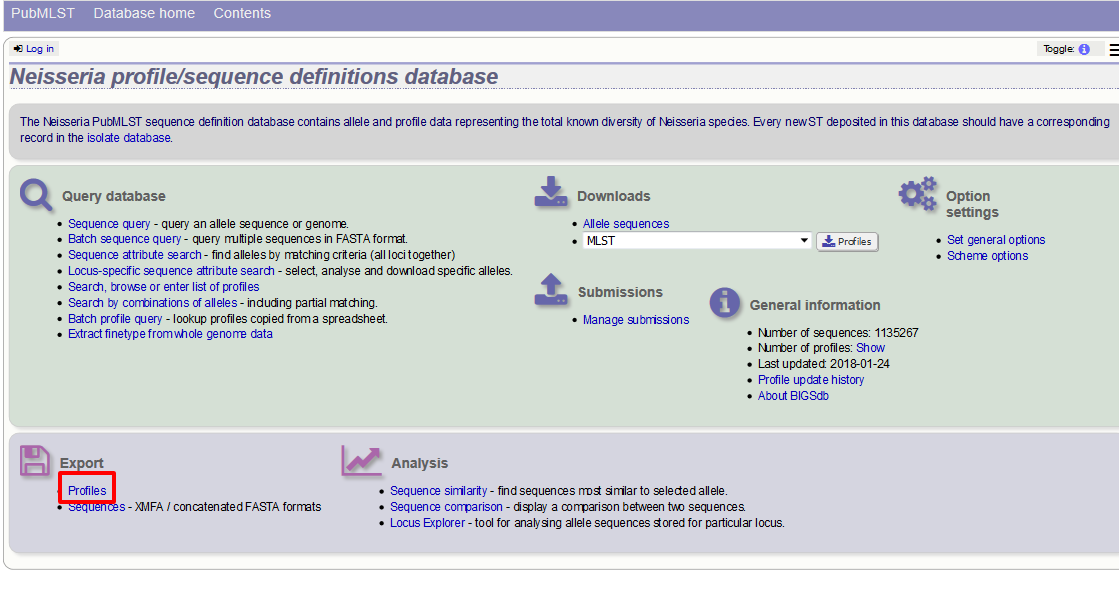
Alternatively, you can access this function by clicking the ‘Profiles’ button in the Export list at the bottom of a results table. Please note that the list of functions here may vary depending on the setup of the database.
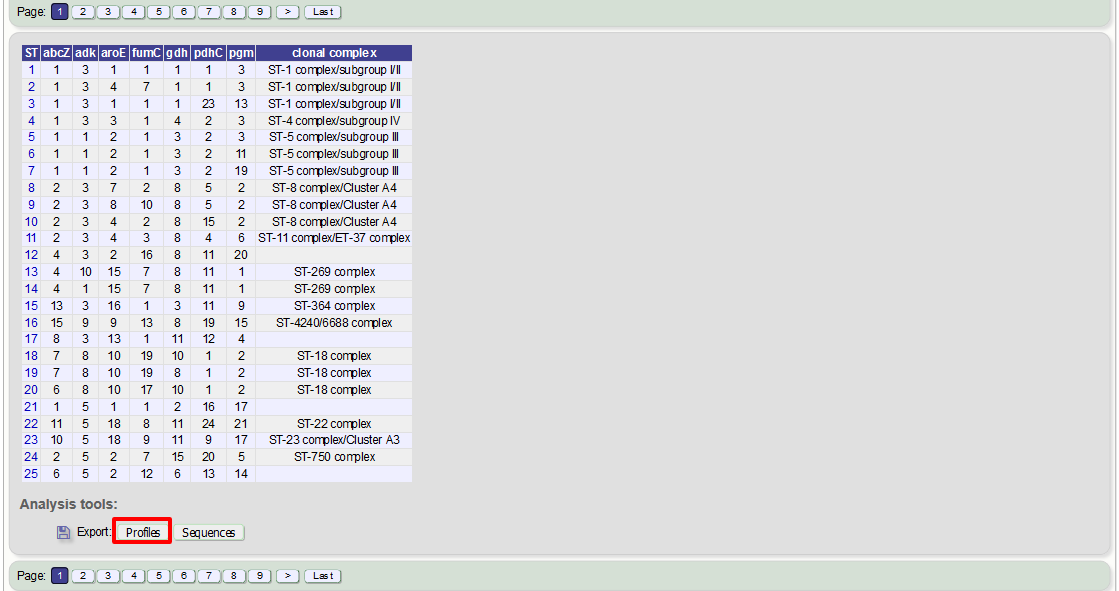
This will take you to a form with a list box in which the identifiers of the profile definitions you wish to include can be entered. Following a query, these values will be pre-entered. If the box is left empty then all profiles will be included.
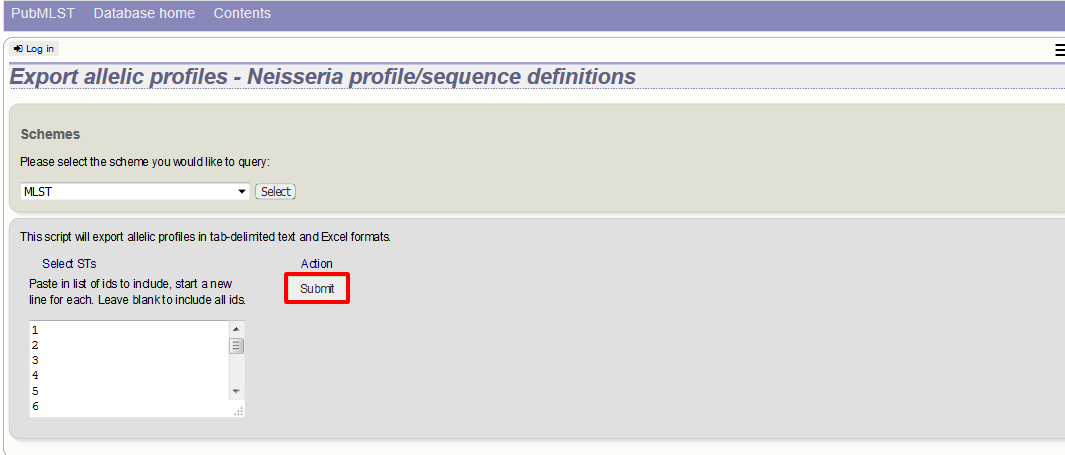
Click submit.
The export job will be submitted to the job queue.
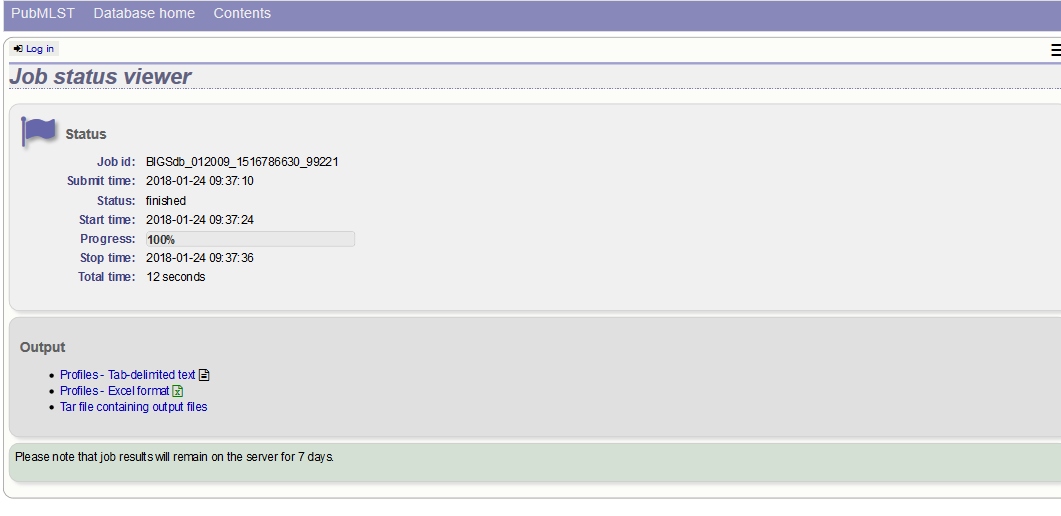
The profiles will be exported in tab-delimited text and Excel formats.
Contig export¶
The contig export plugin can be accessed by clicking the ‘Contigs’ link in the Export section of the contents page of isolate databases.
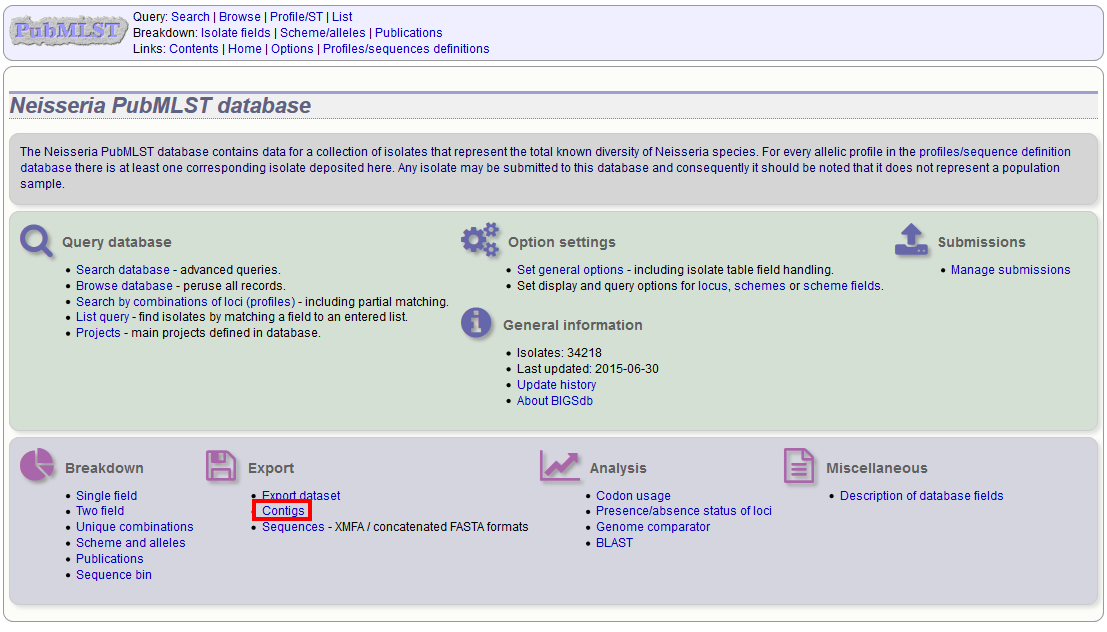
Alternatively, it can be accessed following a query by clicking the ‘Contigs’ button in the Export section at the bottom of the results table.
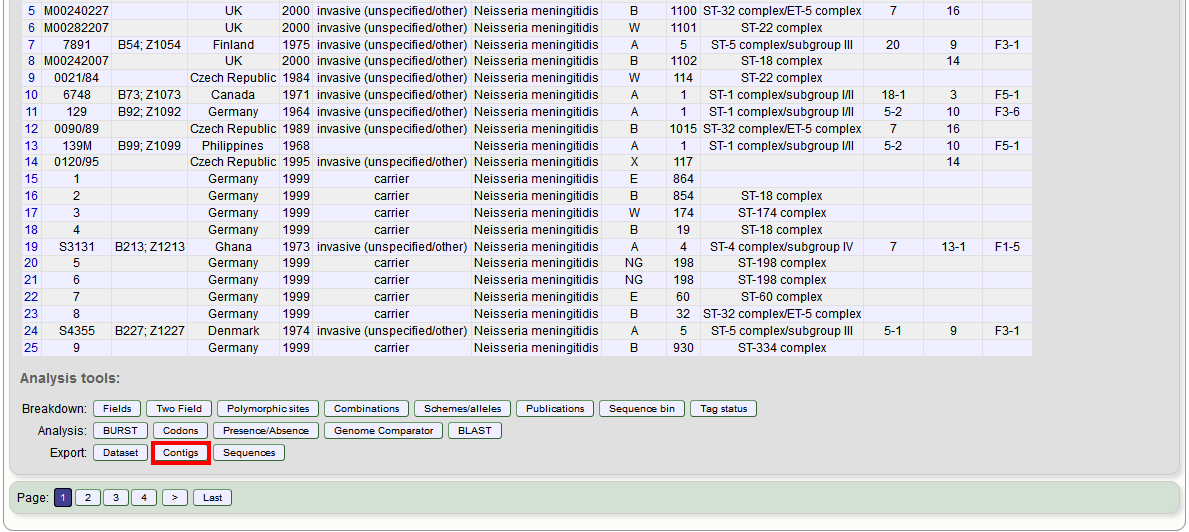
Select the isolates for which you wish to export contig data for. If the export function was accessed following a query, isolates returned in the query will be pre-selected.
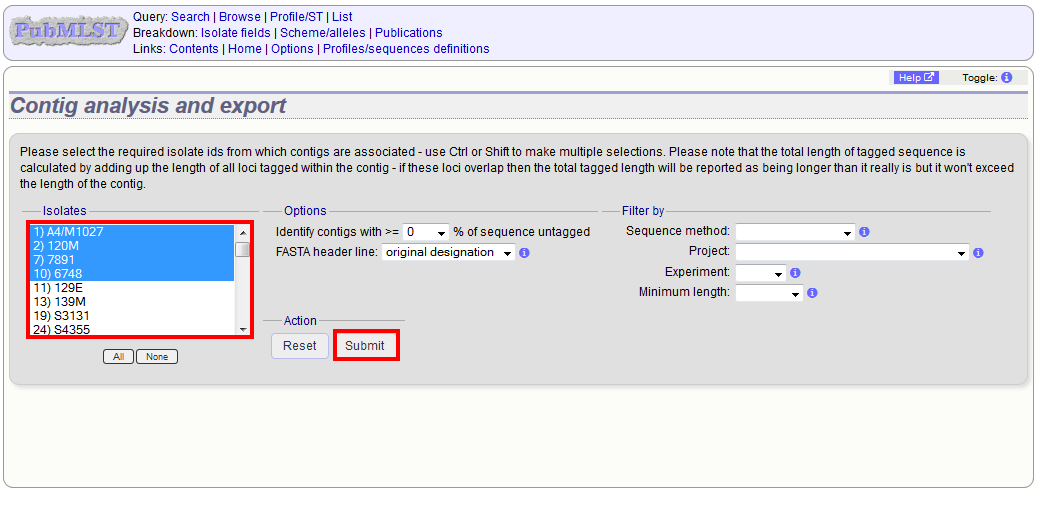
At its simplest, press submit.
A table will be produced with download links. Clicking these will produce the contigs in FASTA format.
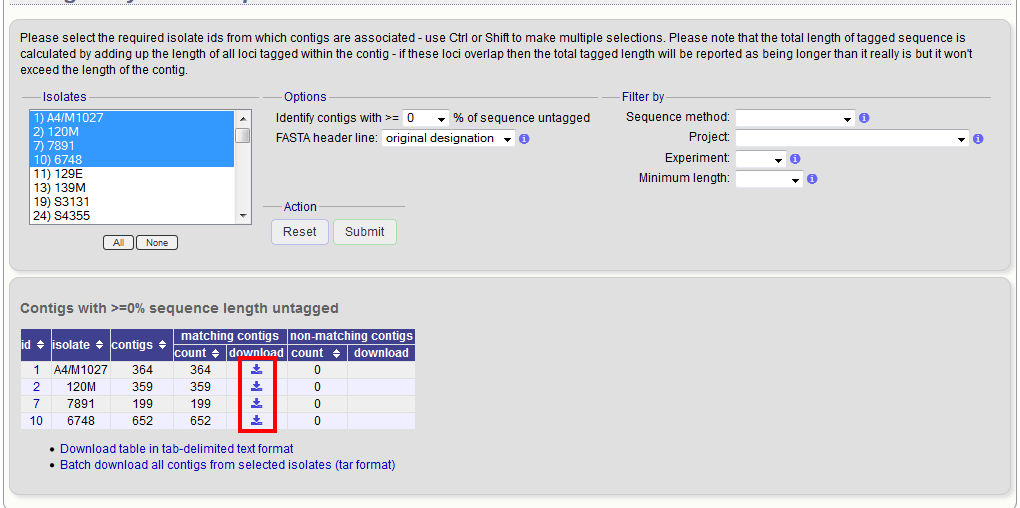
You can also download all the data in a tar file by clicking the ‘Batch download’ link.
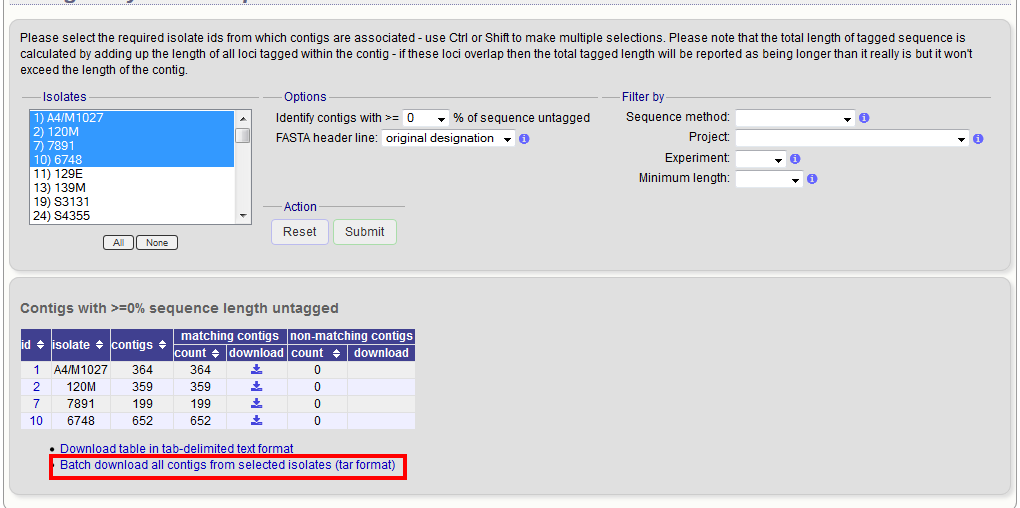
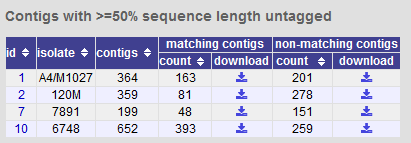
Filtering by tagged status of contigs¶
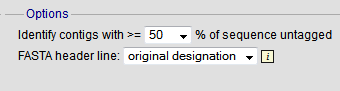
You can also export contigs based on the percentage of the sequence that has been tagged. This is useful to find sequences to target for gene discovery.
In order to export contigs where at least half the sequence has been tagged (and also the remaining contigs in a separate file), select ‘50’ in the dropdown box for %untagged.
The resulting table has two download links for each isolate, one for contigs matching the condition, and one for contigs that don’t match.