Querying isolate data
The ‘Search database’ page of an isolate database allows you to also search by combinations of provenance criteria, scheme and locus data, and more.
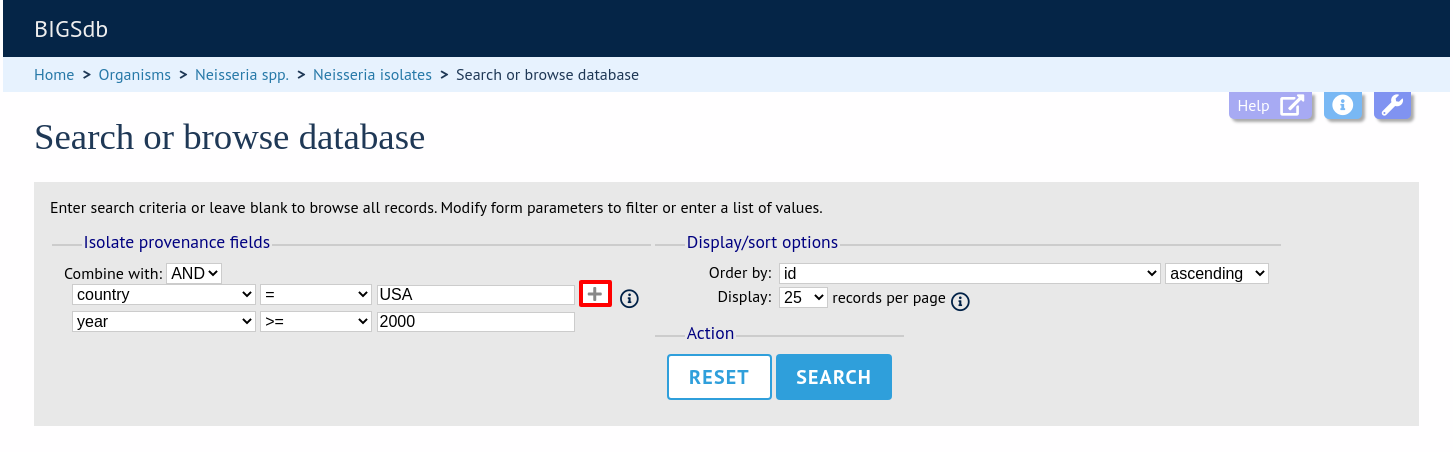
To start with, only one provenance field search box is displayed but more can be added by clicking the ‘+’ button (highlighted). These can be linked together by ‘and’ or ‘or’.
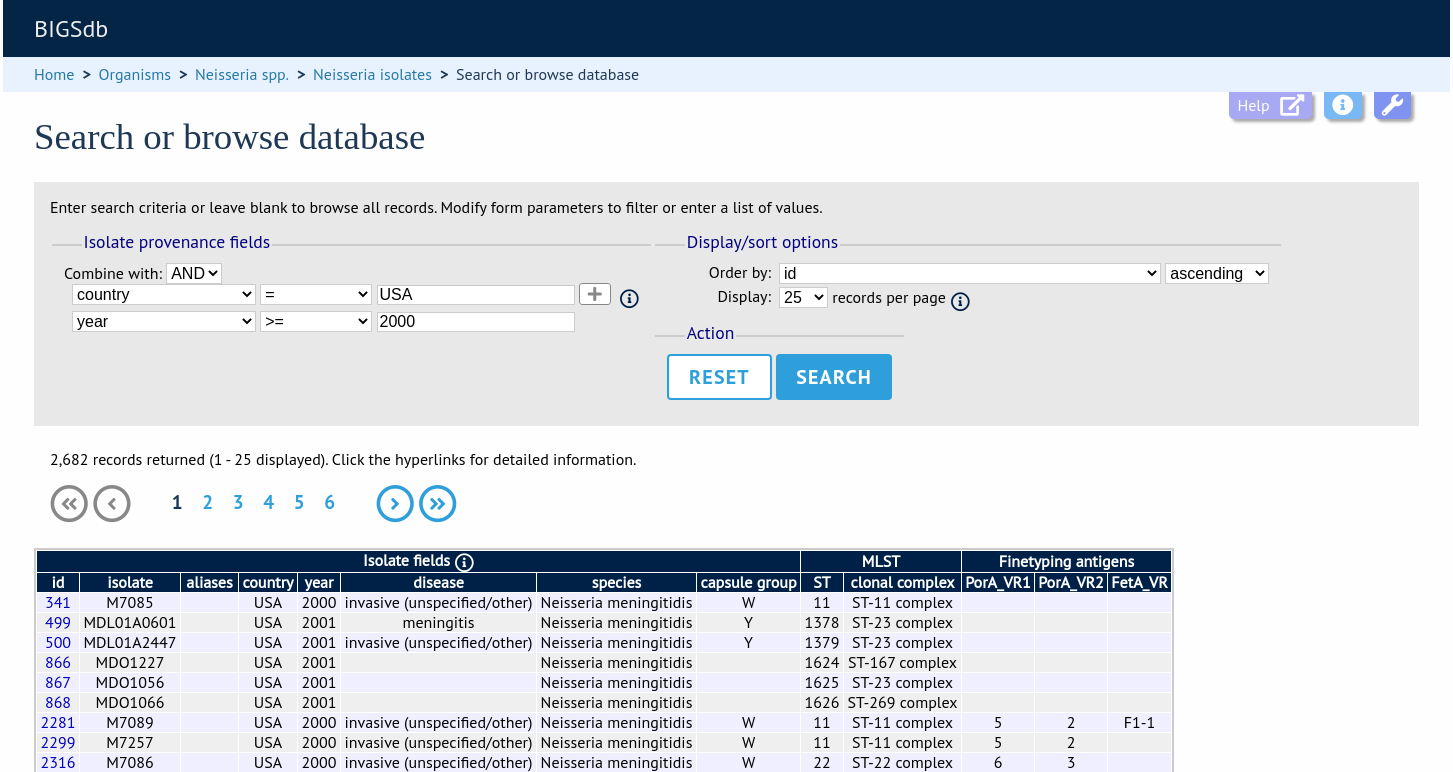
After the search has been submitted, the results will be displayed in a table.
Each field can be queried using standard operators.
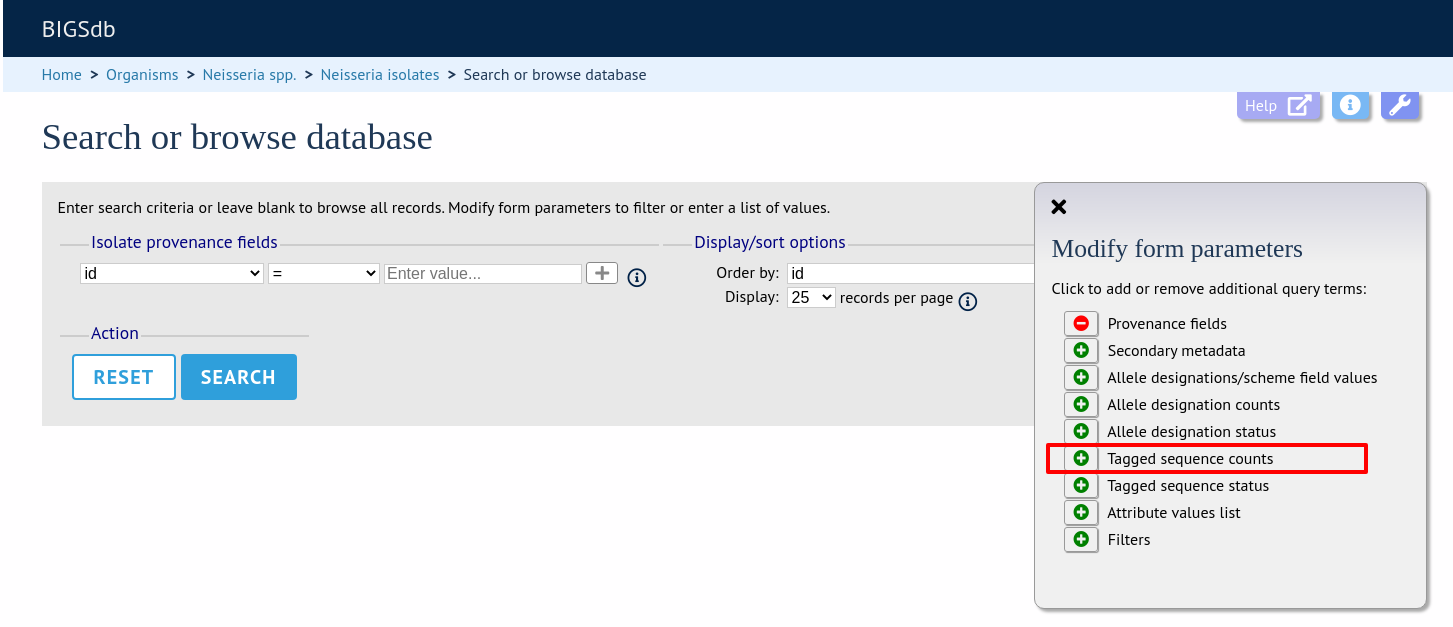
More search features are available by clicking the ‘Modify form options’ tab on the top of the screen.
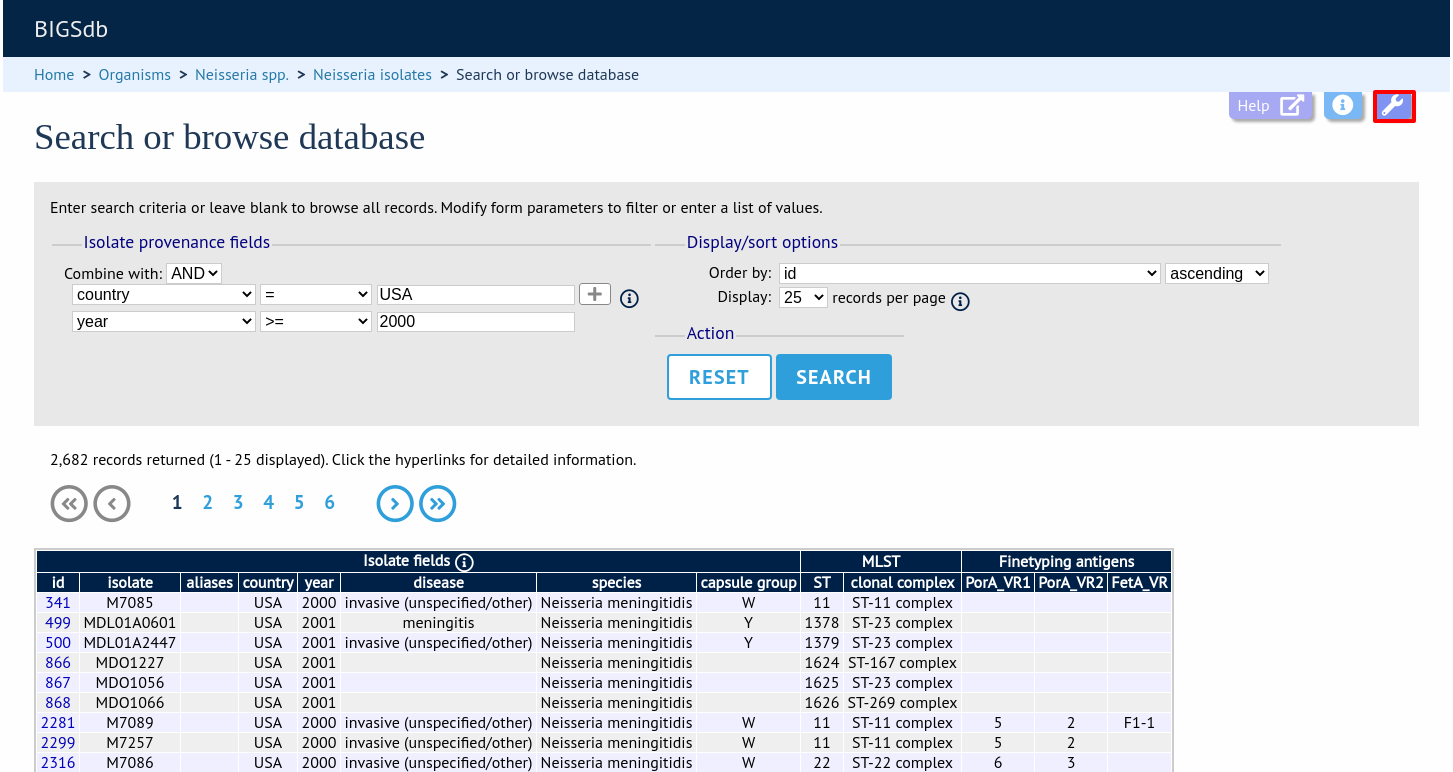
A tab will be displayed. Different options will be available here depending on the database. Queries will be combined from the values entered in all form sections. Possible options are:
Provenance fields
Search by combination of provenance field values, e.g. country, year, sender.
Allele designations/scheme field values
Search by combination of allele designations and/or scheme fields e.g. ST, clonal complex information.
Allele designation status
Search by whether allele designation status is confirmed or provisional.
Tagged sequence status
Search by whether tagged sequence data is available for a locus. You can also search by sequence flags.
Attribute values list
Enter a list of values for any provenance field, locus, or scheme field.
Filters
Various filters may be available, including
Publications
Projects
MLST profile completion status
Clonal complex
Sequence bin size
Inclusion/exclusion of old versions
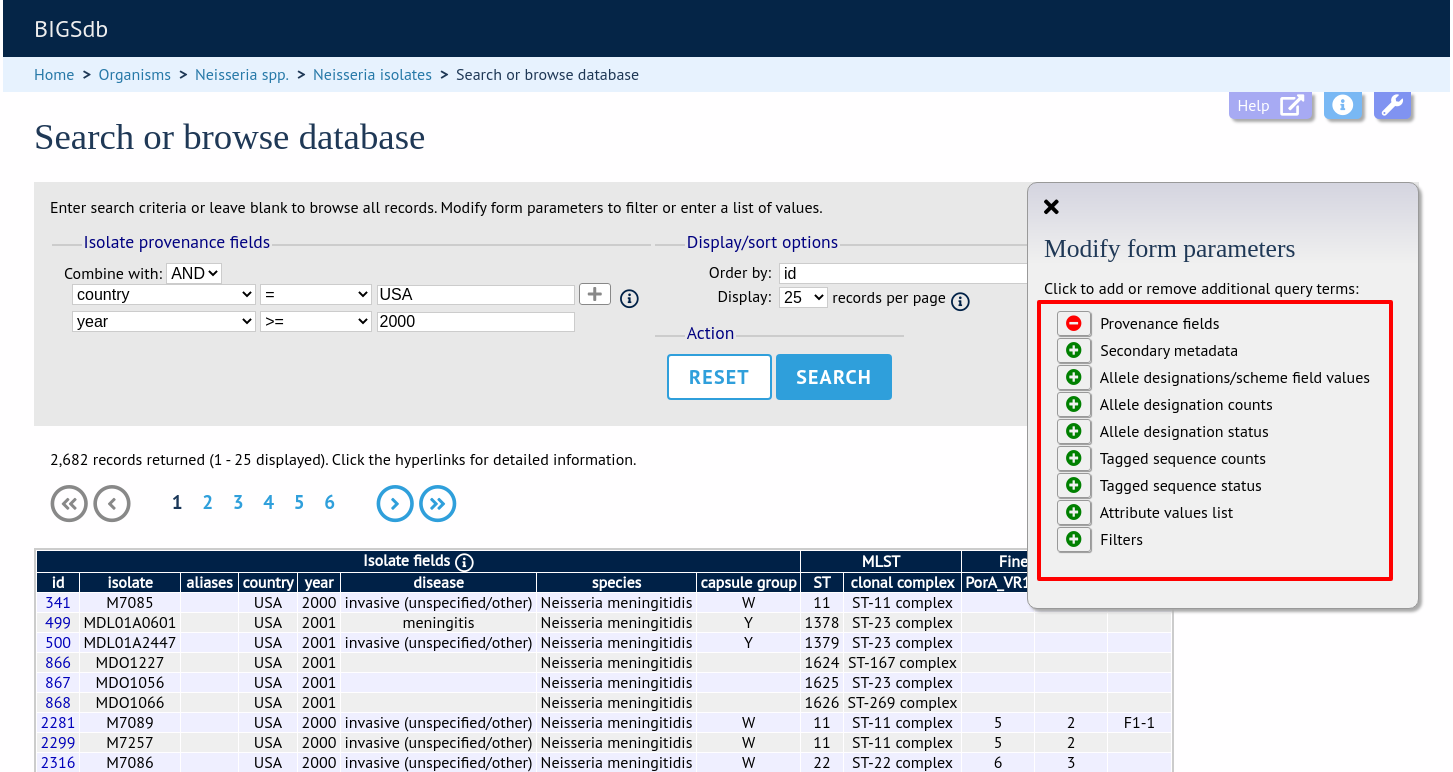
If the interface is modified, a button to save options becomes available within the tab. If this is clicked, the modified form will be displayed the next time you go to the query page.
Query by allele designation/scheme field
Queries can be combined with allele designation/scheme field values.
Make sure that the allele designation/scheme field values fieldset is displayed by selecting it in the ‘Modify form options’ tab.
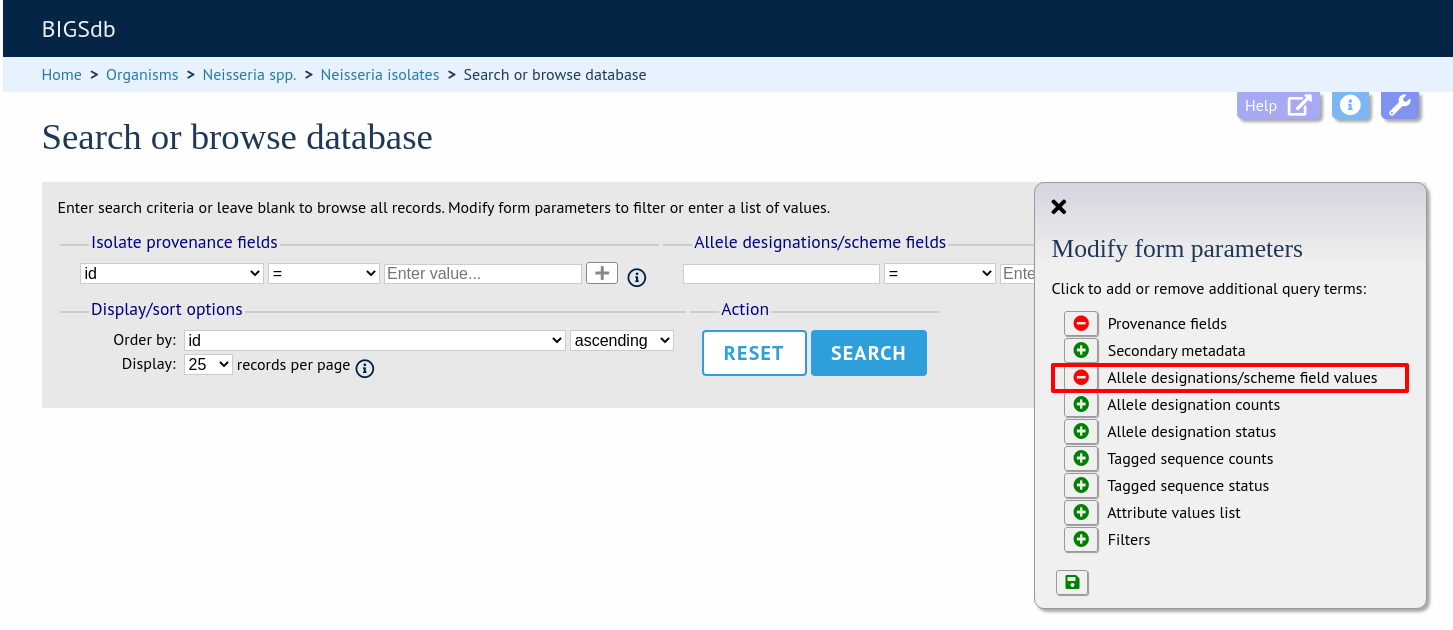
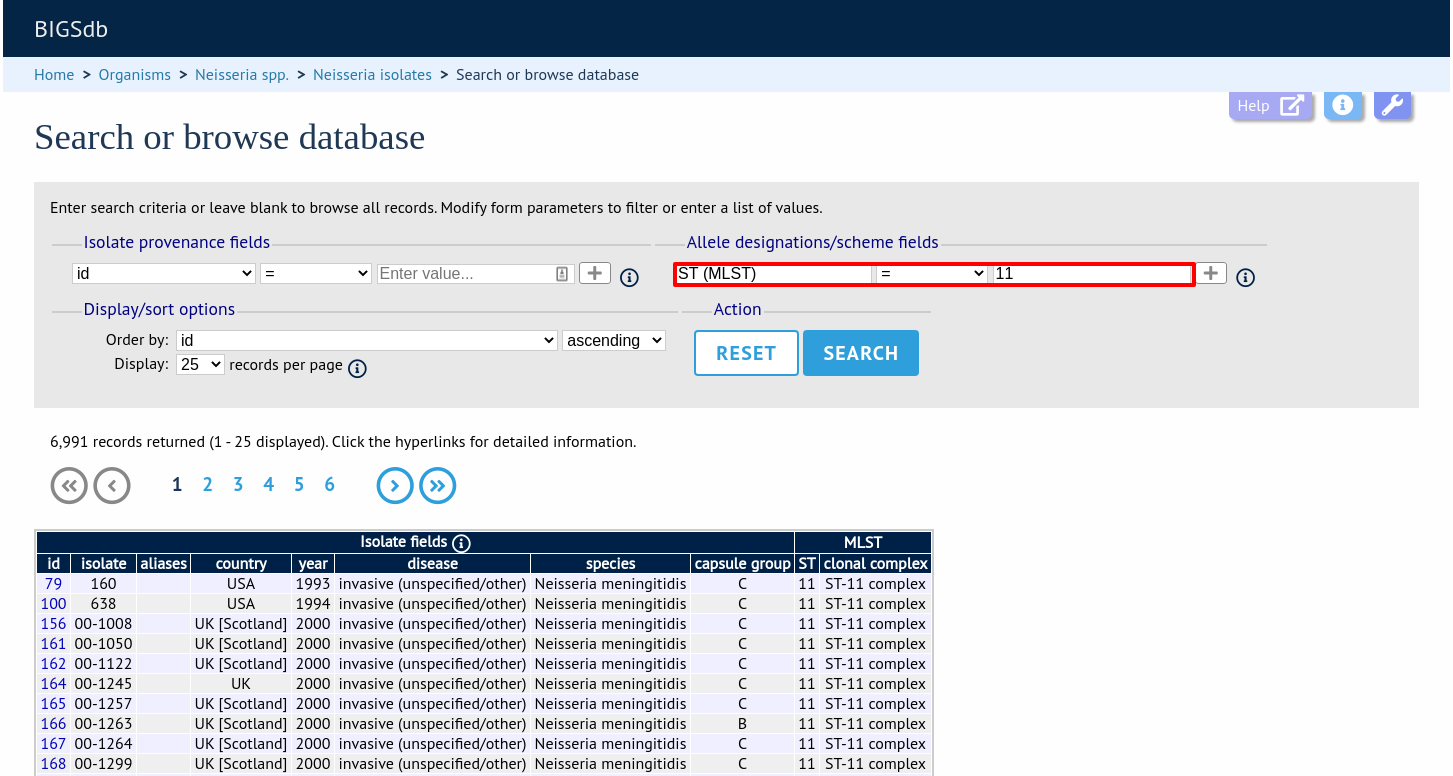
Designations can be queried using standard operators.
Additional search terms can be combined using the ‘+’ button.
Add your search terms and click ‘Submit’. Allele designation/scheme field queries will be combined with terms entered in other sections.
Query by allele designation count
Queries can be combined with counts of the total number of designations or for individual loci.
Make sure that the allele designation counts fieldset is displayed by selecting it in the ‘Modify form options’ tab.
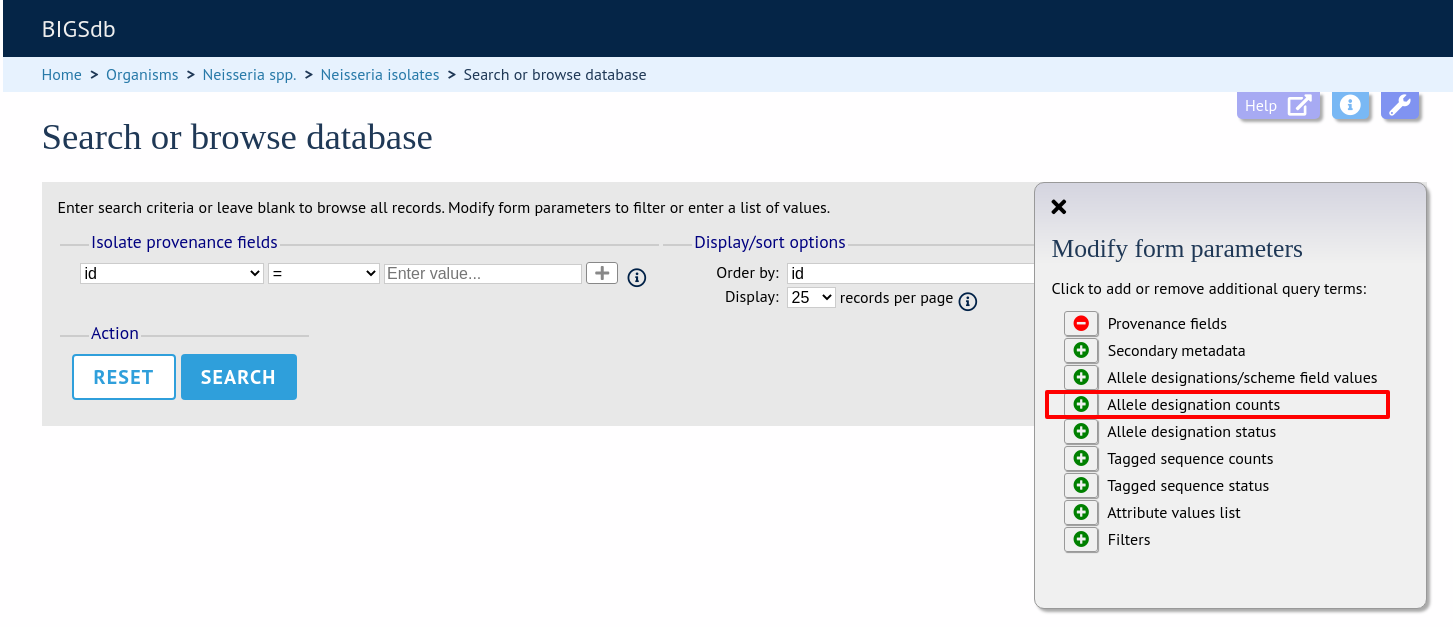
For example, to find all isolates that have designations at >1000 loci, select ‘total designations > 1000’, then click ‘Submit’.
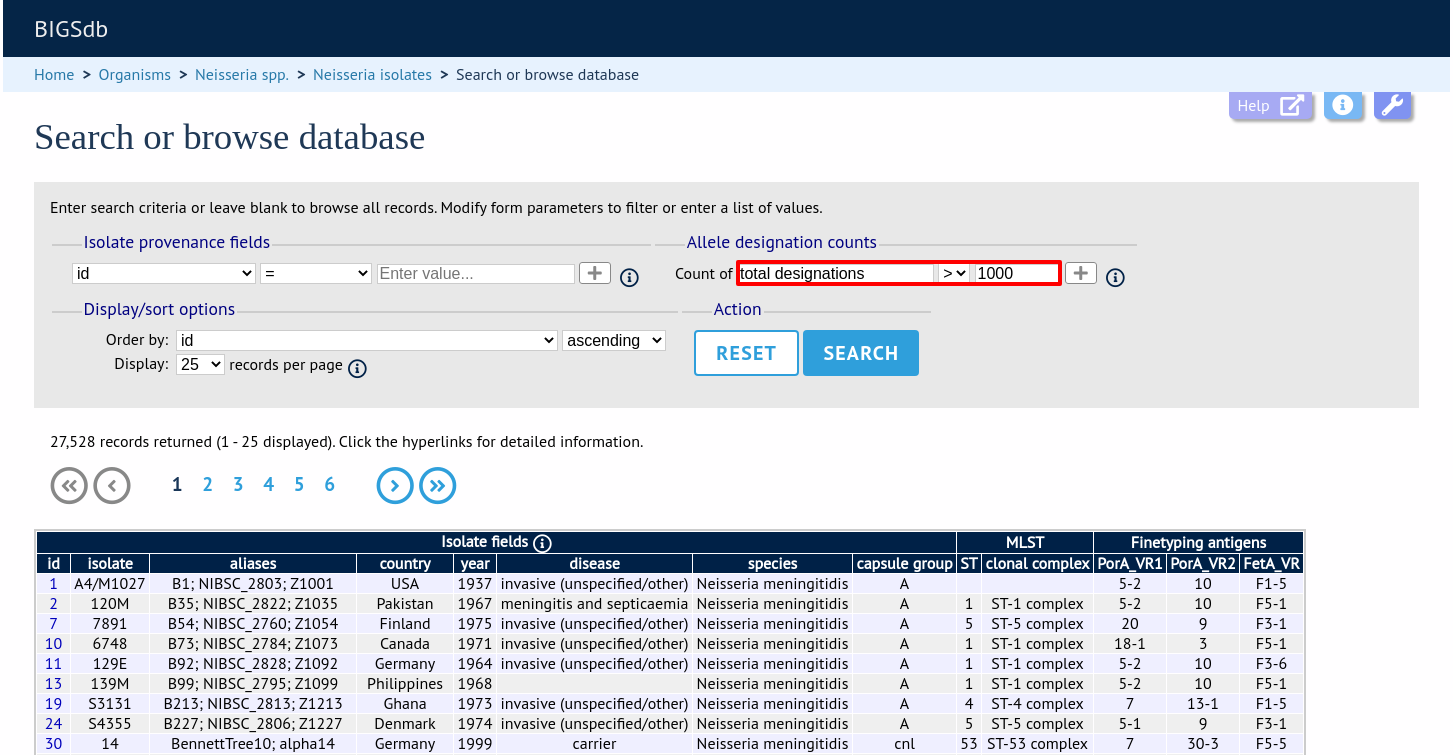
You can also search for isolates where any isolate has a particular number of designations. Use the term ‘any locus’ to do this.
Finally, you can search for isolates with a specific number of designations at a specific locus.
Additional search terms can be combined using the ‘+’ button. Designation count queries will be combined with terms entered in other sections.
Note
Searches for ‘all loci’ with counts that include zero, e.g. ‘count of any locus = 0’ or with a ‘<’ operator are not supported. This is because such searches have to identify every isolate for which one or more loci are missing. In databases with thousands of loci this can be a very expensive database query.
Query by allele designation status
Allele designations can be queried based on their status, i.e. whether they are confirmed or provisional. Queries will be combined from the values entered in all form sections.
Make sure that the allele designation staus fieldset is displayed by selecting it in the ‘Modify form options’ tab.
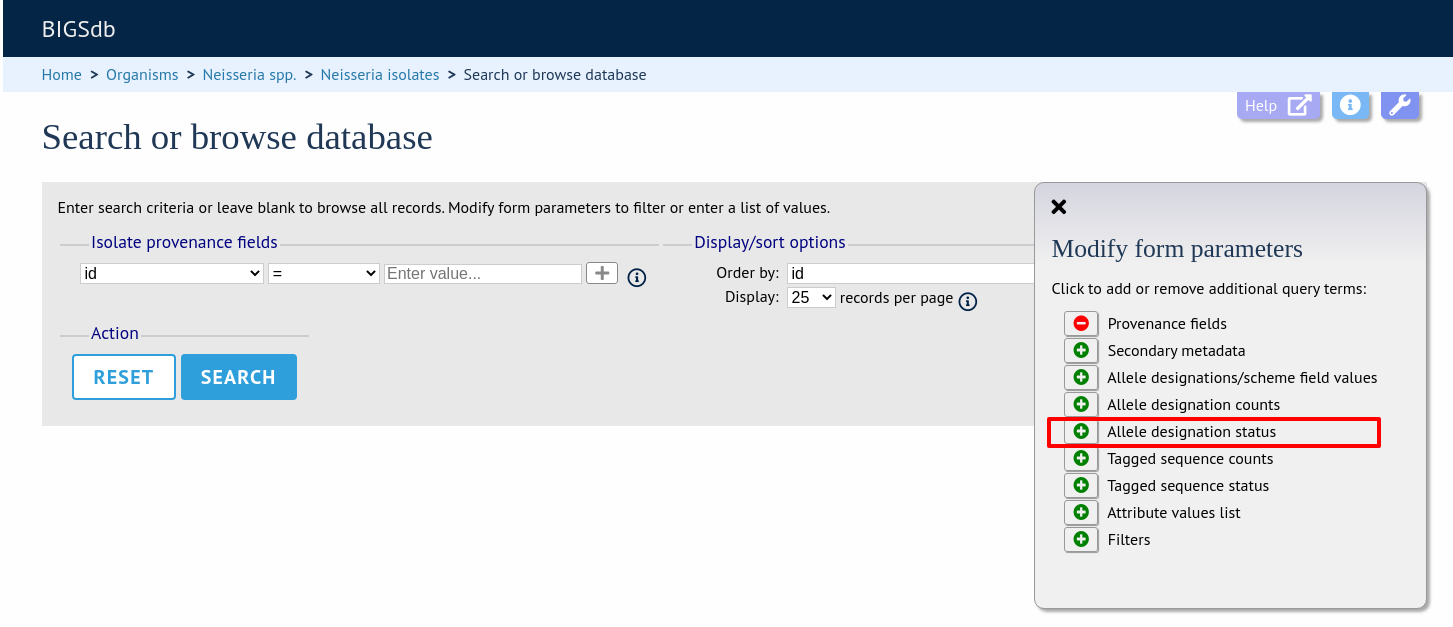
Select a locus from the dropdown box and either ‘provisional’ or ‘confirmed’. Additional query fields can be displayed by clicking the ‘+’ button. Click ‘Submit’.
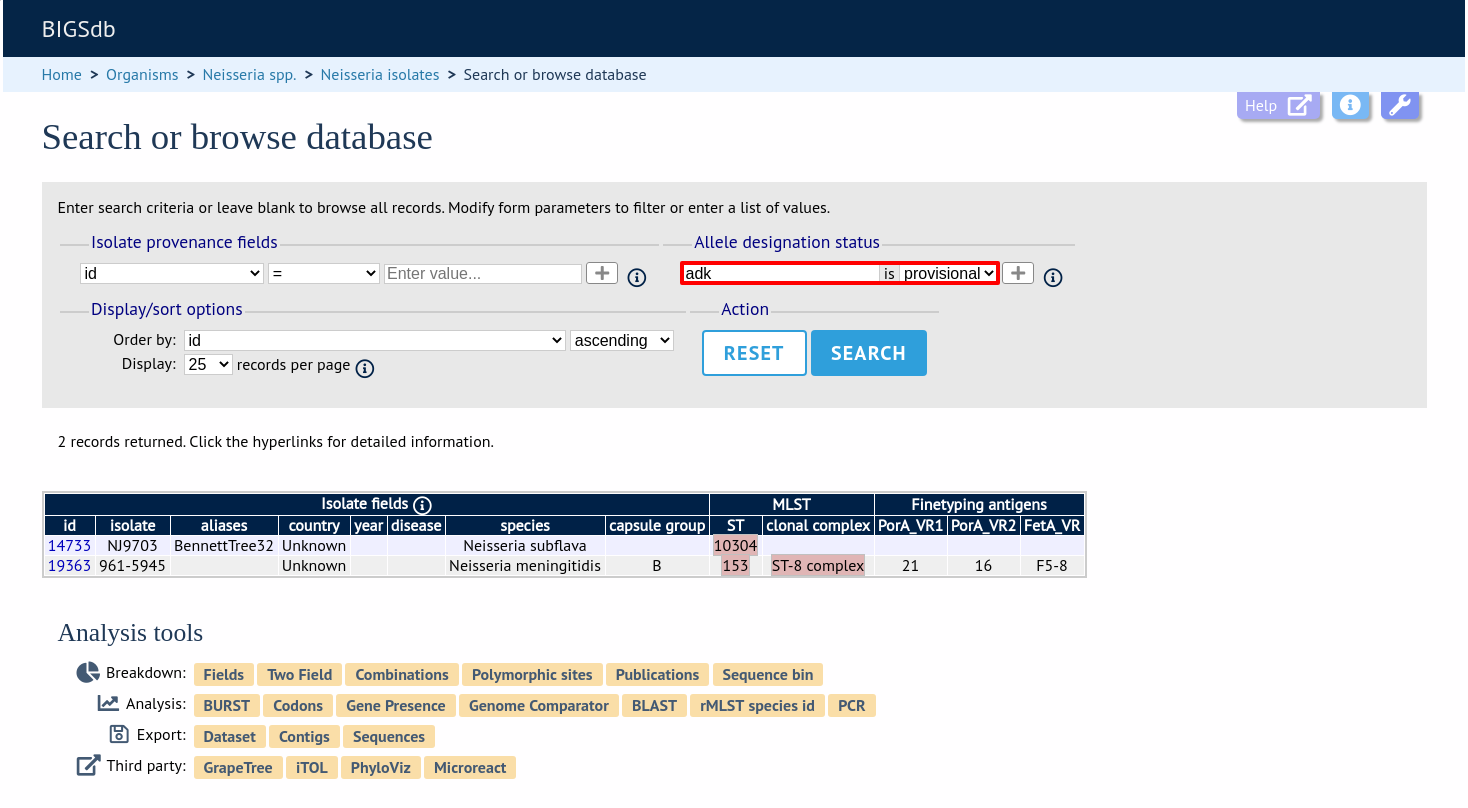
Provisional allele designations are marked within the results tables with a pink background. Any scheme field designations that depend on the allele in question, e.g. a MLST ST, will also be marked as provisional.
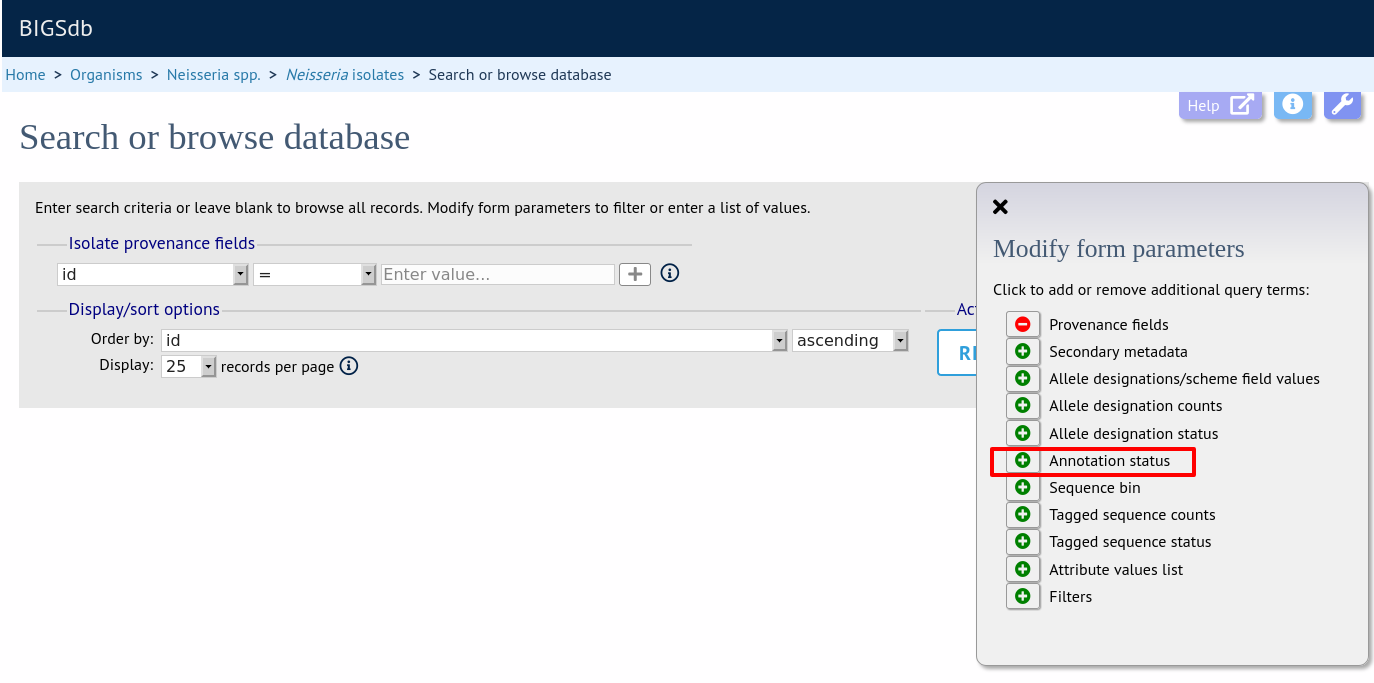
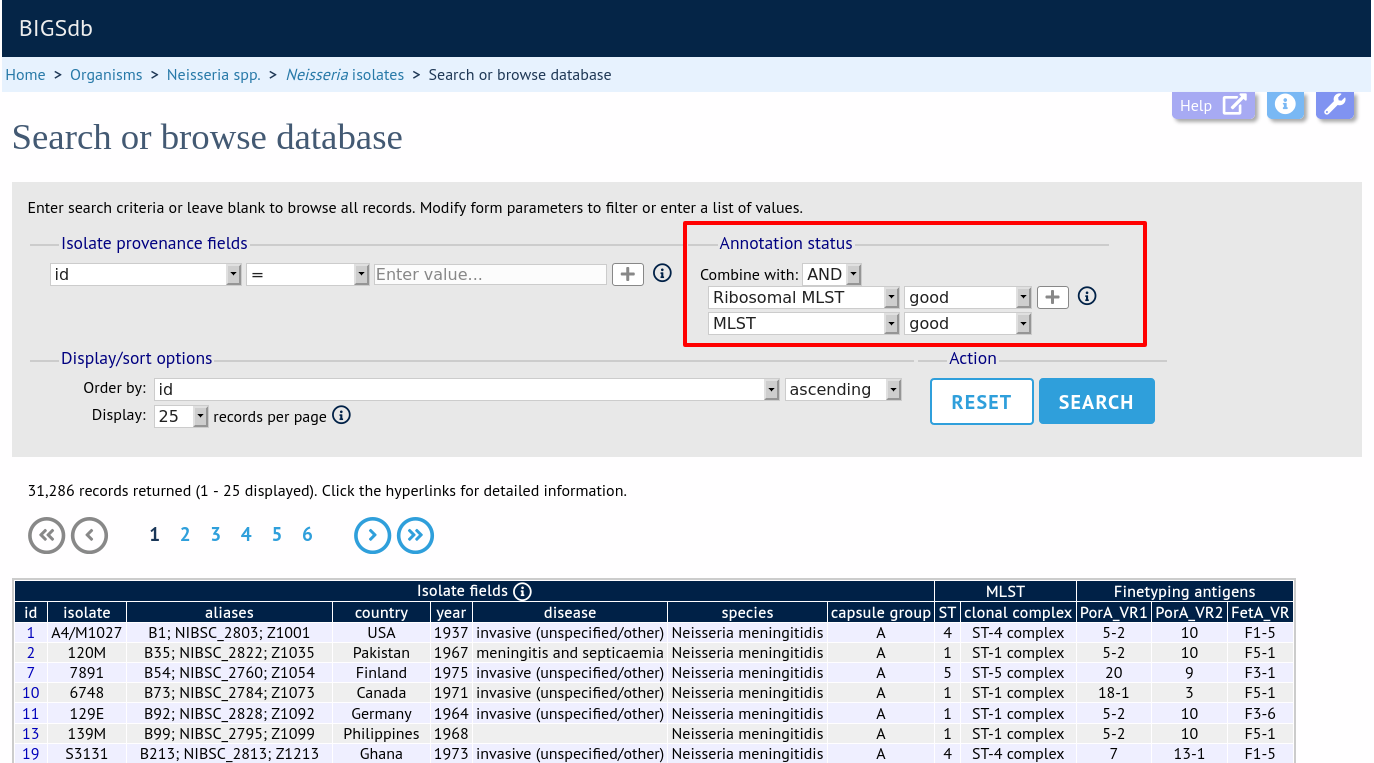
Query by annotation status
Isolates can be queried by the annotation status of particular schemes if these have been set up. The idea is that for a well-annotated record the isolate would be expected to have allele designations for all loci in the scheme. Alternatively, different thresholds for number of loci with allele designations can be set up by the scheme administrator to indicate good or bad quality thresholds.
Make sure that the annotation status fieldset is displayed by selecting it in the ‘Modify form options’ tab.
Additional search terms can be combined using the ‘+’ button. Annotation status queries will be combined with terms entered in other sections.
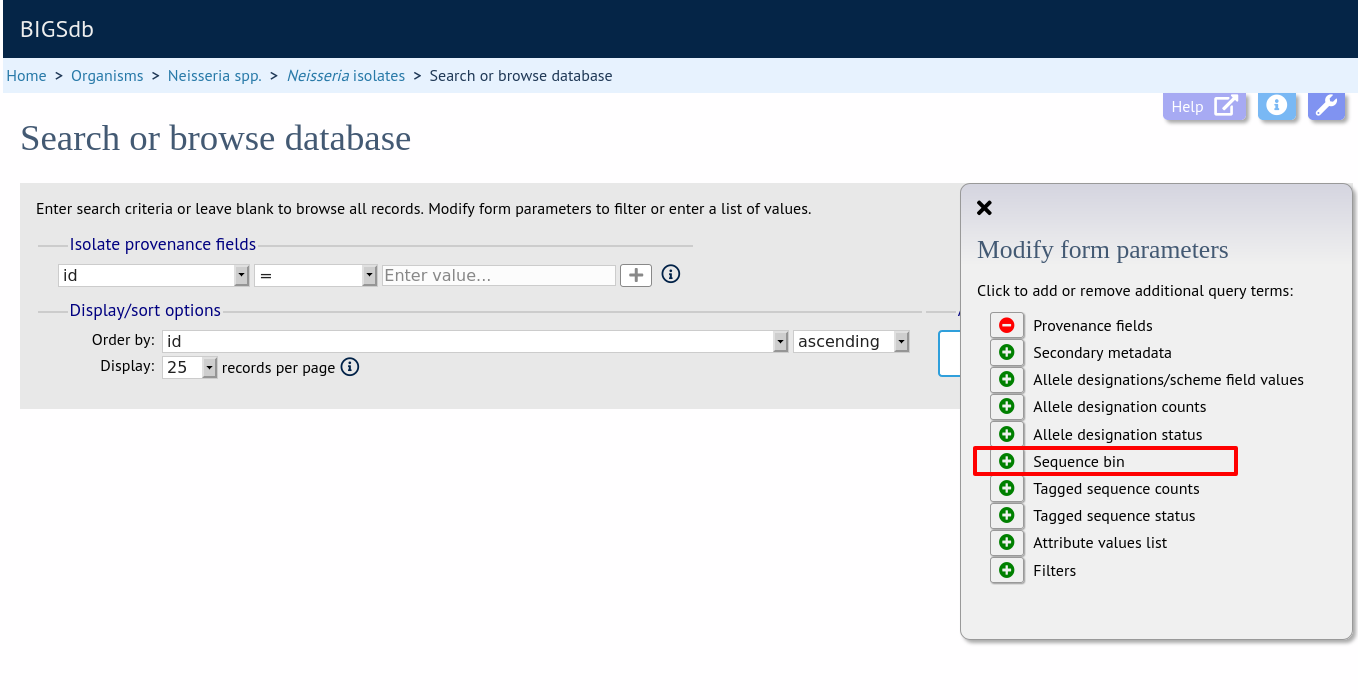
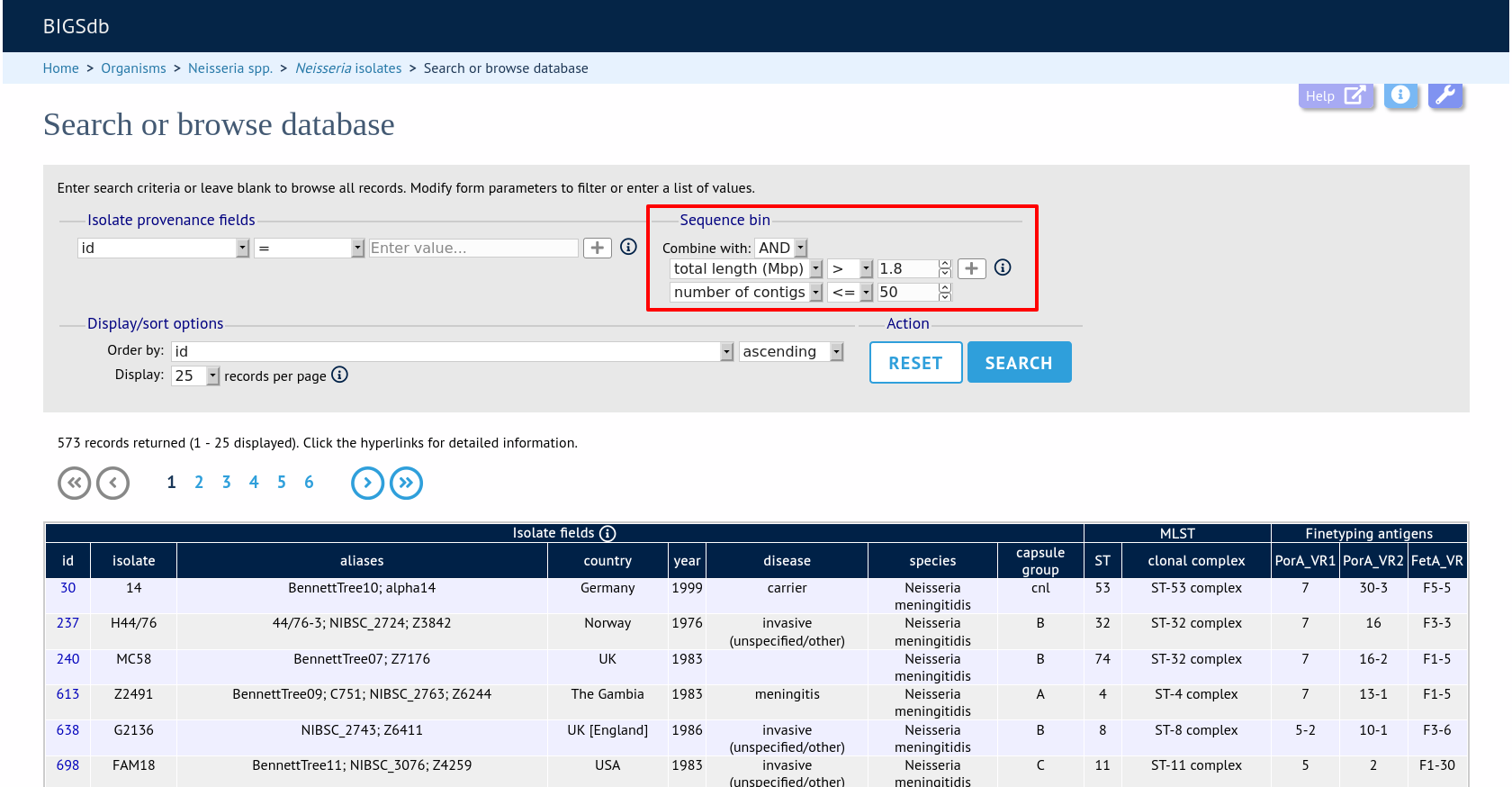
Query by sequence bin size and number of contigs
Isolates can be queried based on the total length of sequences within the sequence bin, the number of contigs, the N50 and/or the L50 values.
Make sure that the sequence bin fieldset is displayed by selecting it in the ‘Modify form options’ tab.
Additional search terms can be combined using the ‘+’ button. Sequence bin queries will be combined with terms entered in other sections.
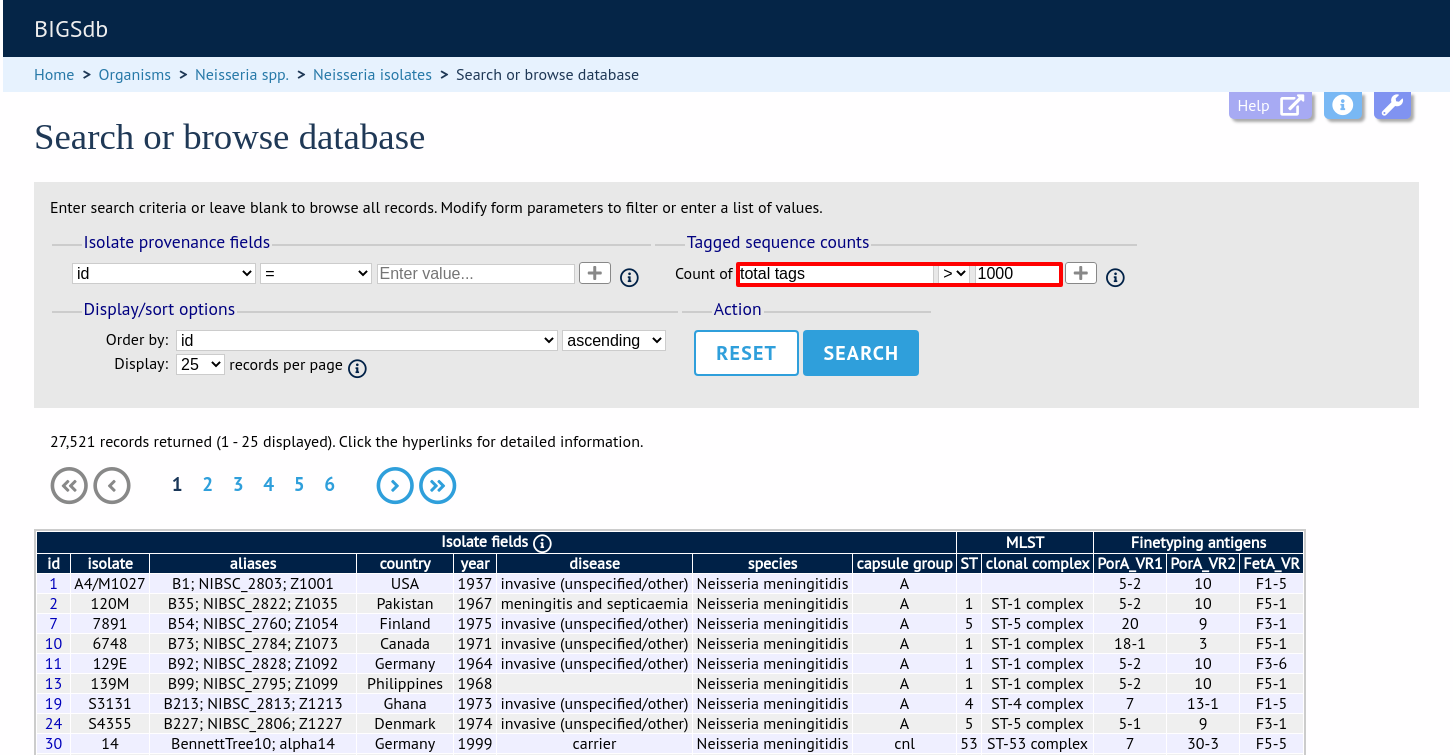
Query by sequence tag count
Queries can be combined with counts of the total number of tags or for individual loci.
Make sure that the tagged sequence counts fieldset is displayed by selecting it in the ‘Modify form options’ tab.
For example, to find all isolates that have sequence tags at >1000 loci, select ‘total tags > 1000’, then click ‘Submit’.
You can also search for isolates where any isolate has a particular number of sequence tags. Use the term ‘any locus’ to do this.
Finally, you can search for isolates with a specific number of tags at a specific locus.
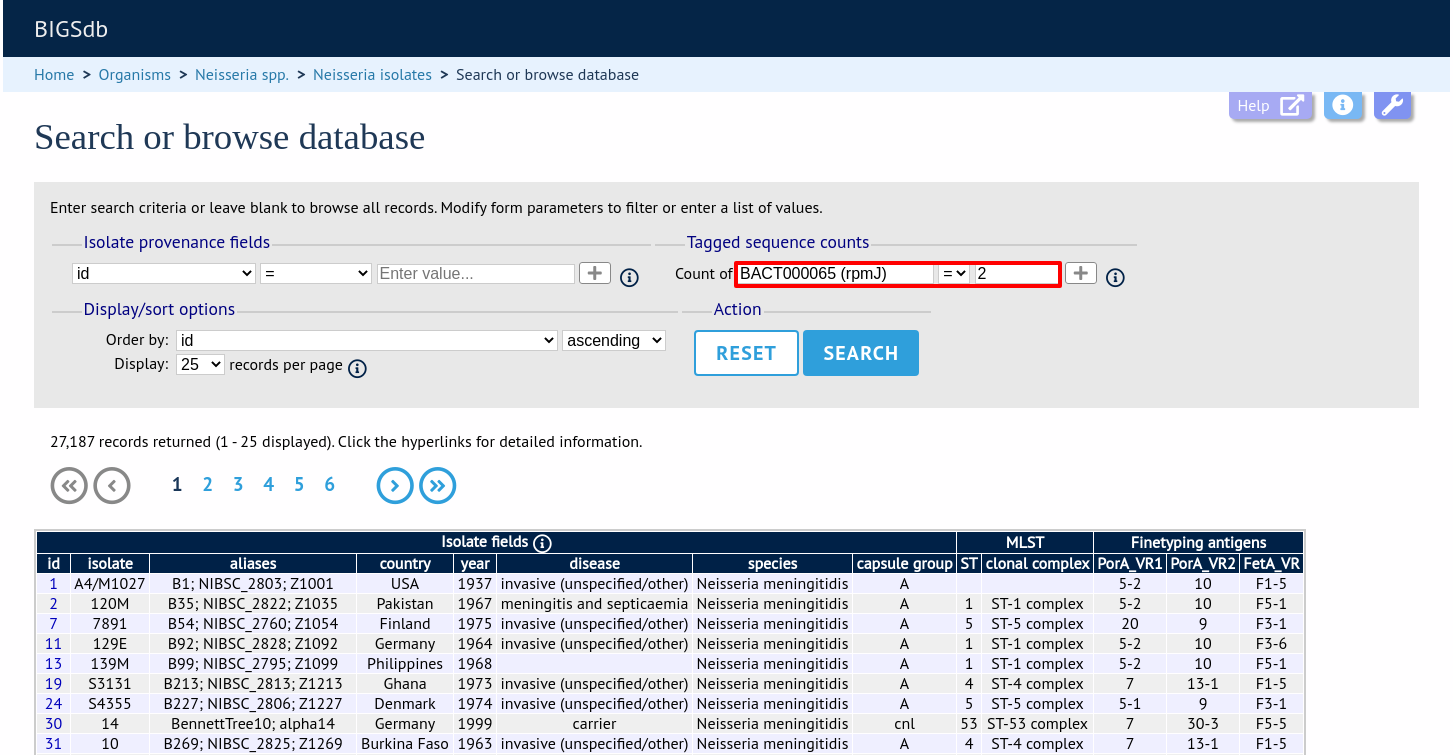
Additional search terms can be combined using the ‘+’ button. Sequence tag count queries will be combined with terms entered in other sections.
Note
Searches for ‘all loci’ with counts that include zero, e.g. ‘count of any locus = 0’ or with a ‘<’ operator are not supported. This is because such searches have to identify every isolate for which one or more loci are not tagged. In databases with thousands of loci this can be a very expensive database query.
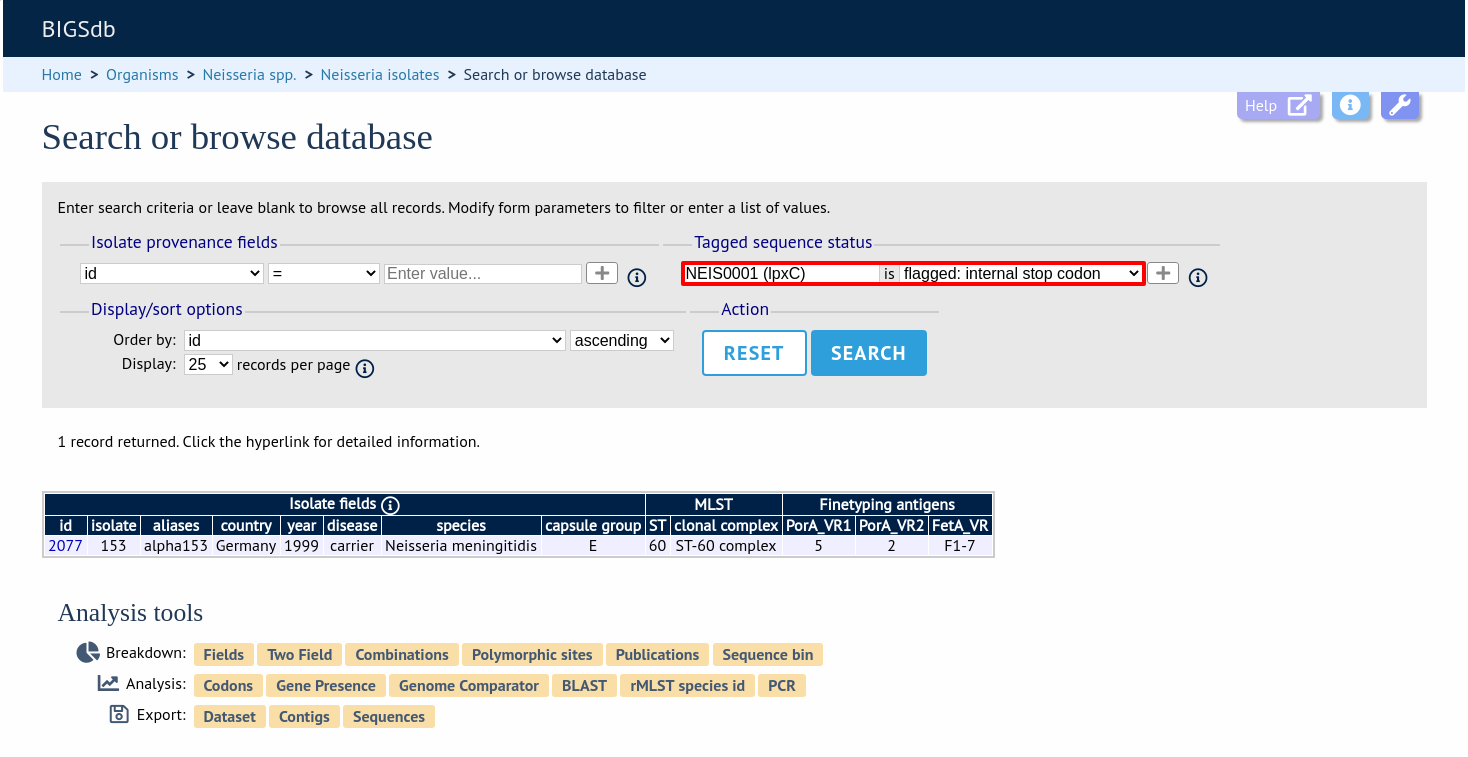
Query by tagged sequence status
Sequence tags identify the region of a contig within an isolate’s sequence bin entries that correspond to a particular locus. The presence or absence of these tags can be queried as can whether or not the sequence has an a flag associated with. These flags designate specific characteristics of the sequences. Queries will be combined from the values entered in all form sections.
Make sure that the tagged sequences status fieldset is displayed by selecting it in the ‘Modify form options’ tab.
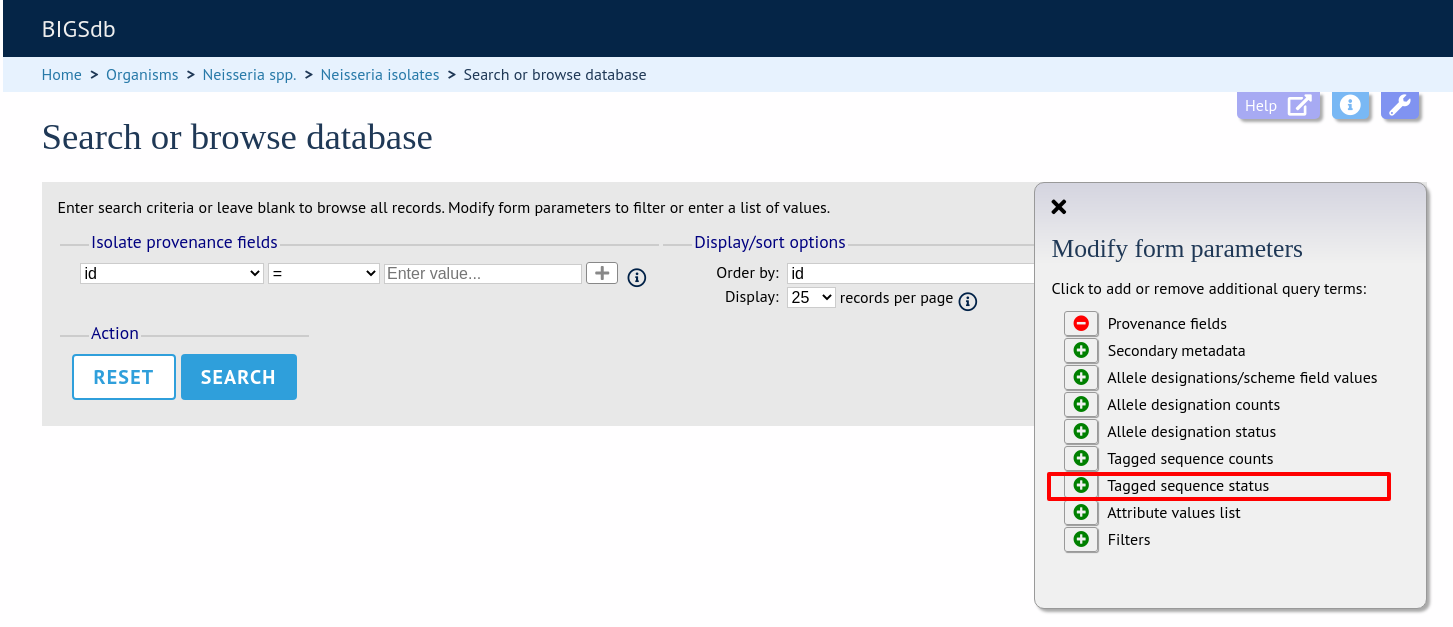
Select a specific locus in the dropdown box (or alternatively ‘any locus’) and a status. Available status values are:
untagged
The locus has not been tagged within the sequence bin.
tagged
The locus has been tagged within the sequence bin.
complete
The locus sequence is complete.
incomplete
The locus sequence is incomplete - normally because it continues beyond the end of a contig.
flagged: any
The sequence for the locus has a flag set.
flagged: none
The sequence for the locus does not have a flag set.
flagged: <specific flag>
The sequence for the locus has the specific flag chosen.
See also
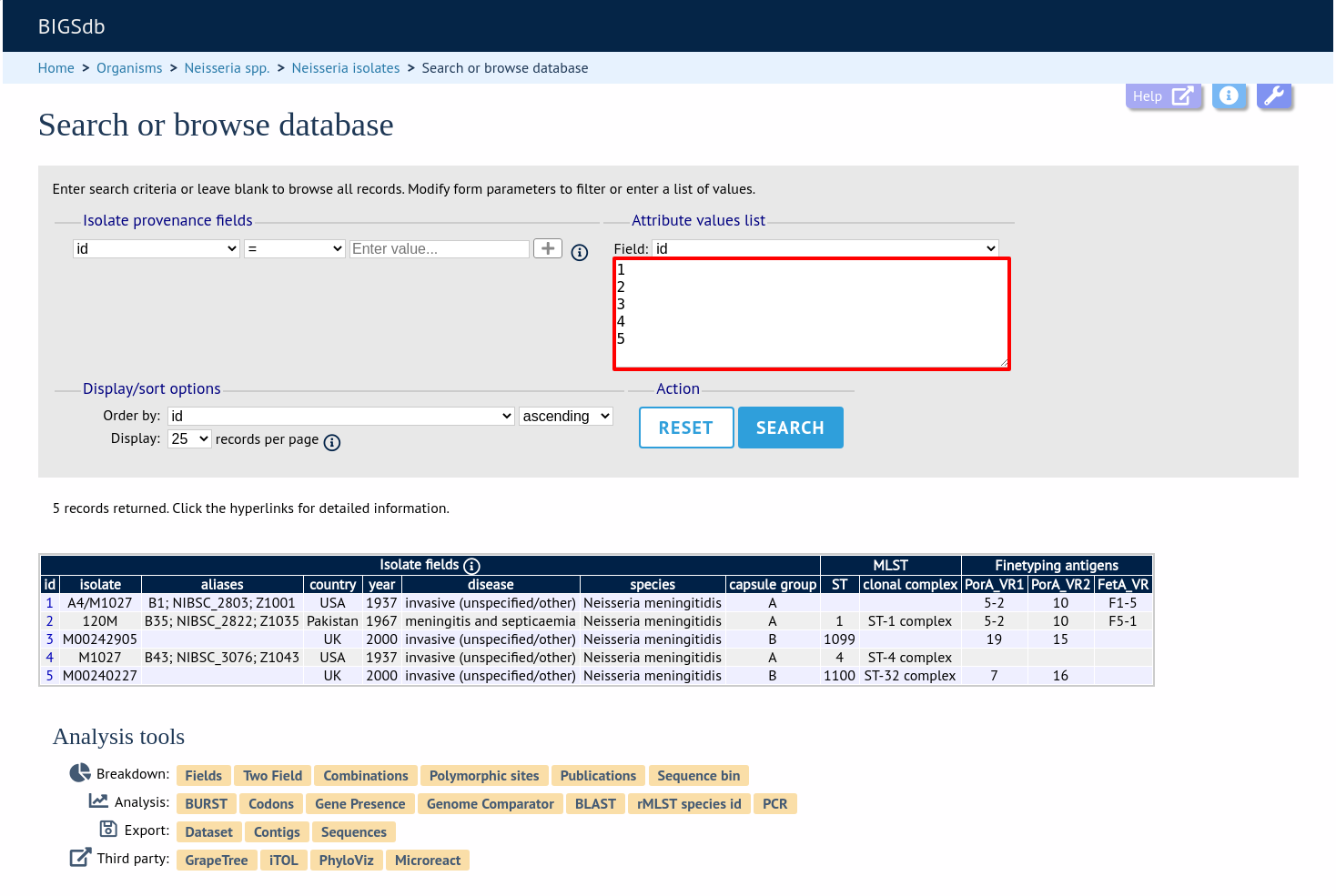
Query by list of attributes
The query form can be modified with a list box in to which a list of values for a chosen attribute can be entered - this could be a list of ids, isolate names, alleles or scheme fields. This list will be combined with any other criteria or filter used on the page.
If the list box is not shown, add it by selecting it in the ‘Modify form options’ tab.
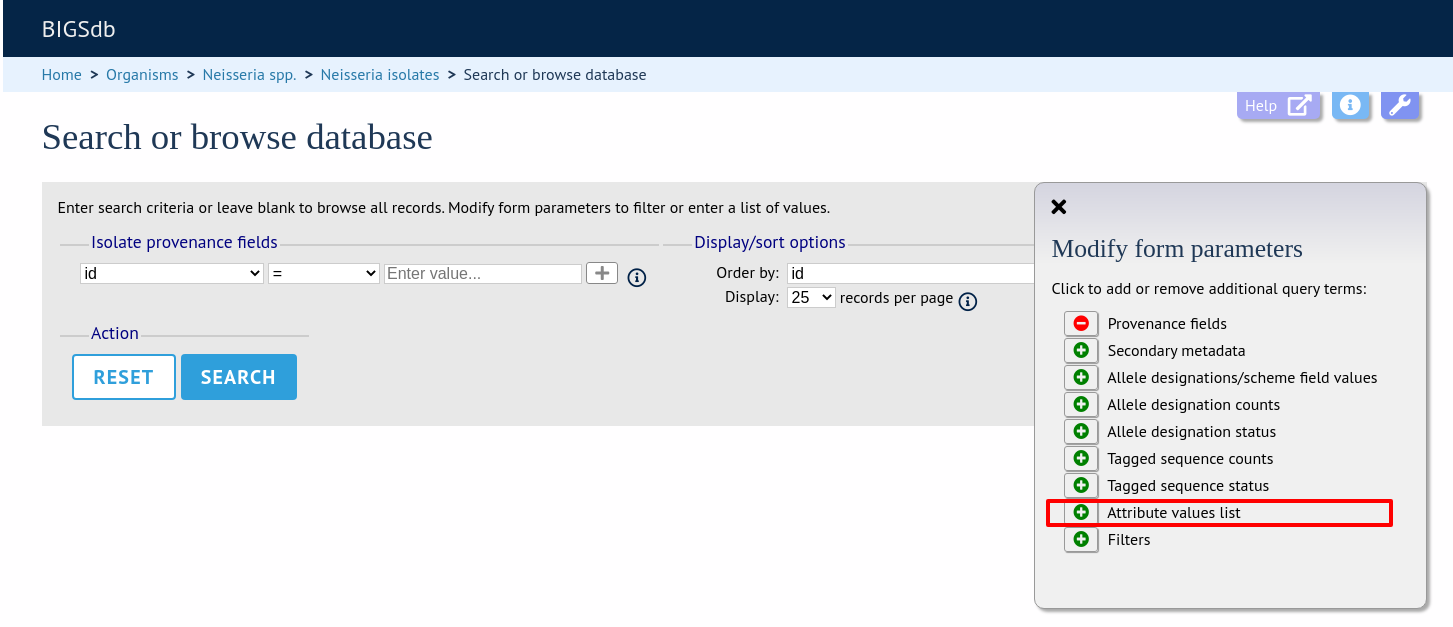
Select the attribute to query and enter a list of values.
Query filters
There are various filters that can additionally be applied to queries, or the filters can be applied solely on their own so that they filter the entire database.
Make sure that the filters fieldset is displayed by selecting it in the ‘Modify form options’ tab.
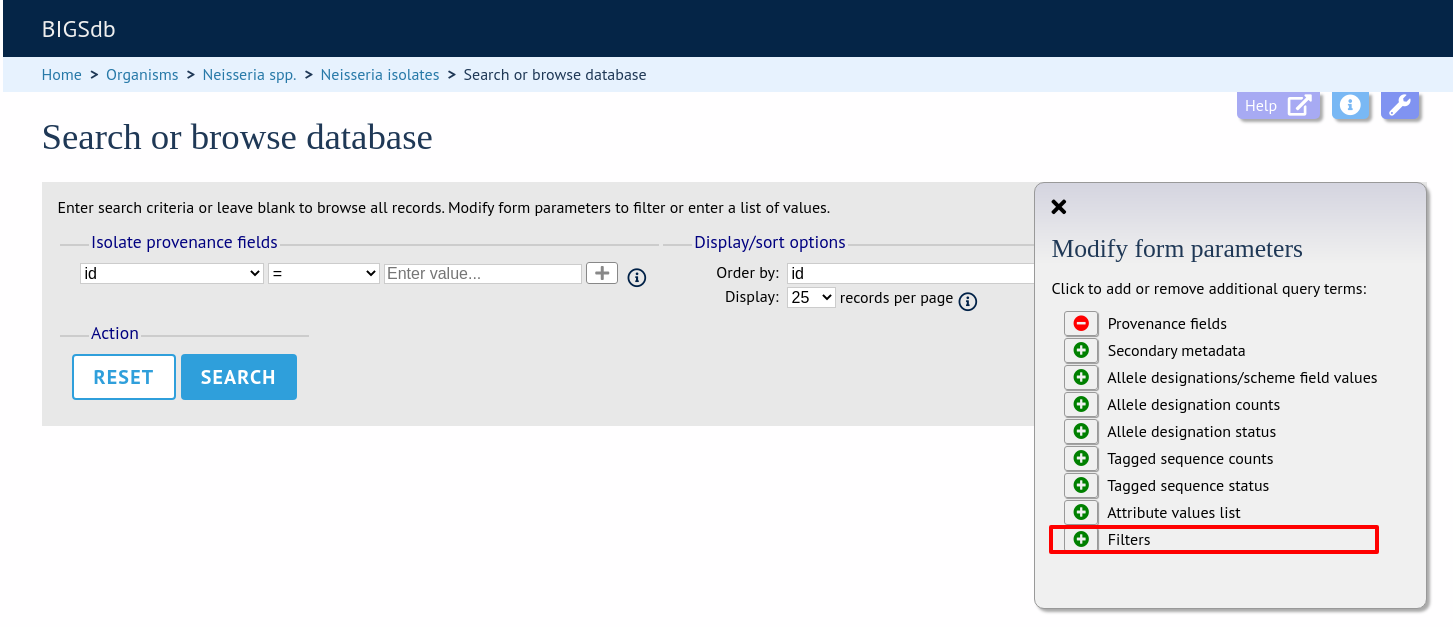
The filters displayed will depend on the database and what has been defined within it. Common filters are:
Publication - Select one or more publication that has been linked to isolate records.
Project - Select one or more project that isolates belong to.
Profile completion - This is commonly displayed for MLST schemes. Available options are:
complete - All loci of the scheme have alleles designated.
incomplete - One or more loci have not yet been designated.
partial - The scheme is incomplete, but at least one locus has an allele designated.
started - At least one locus has an allele designated. The scheme mat be complete or partial.
not started - The scheme has no loci with alleles designated.
Provenance fields - Dropdown list boxes of values for specific provenance fields may be present if set for the database. Users can choose to add additional filters.
Old record versions - Checkbox which, if selected, will include all record versions in a query.
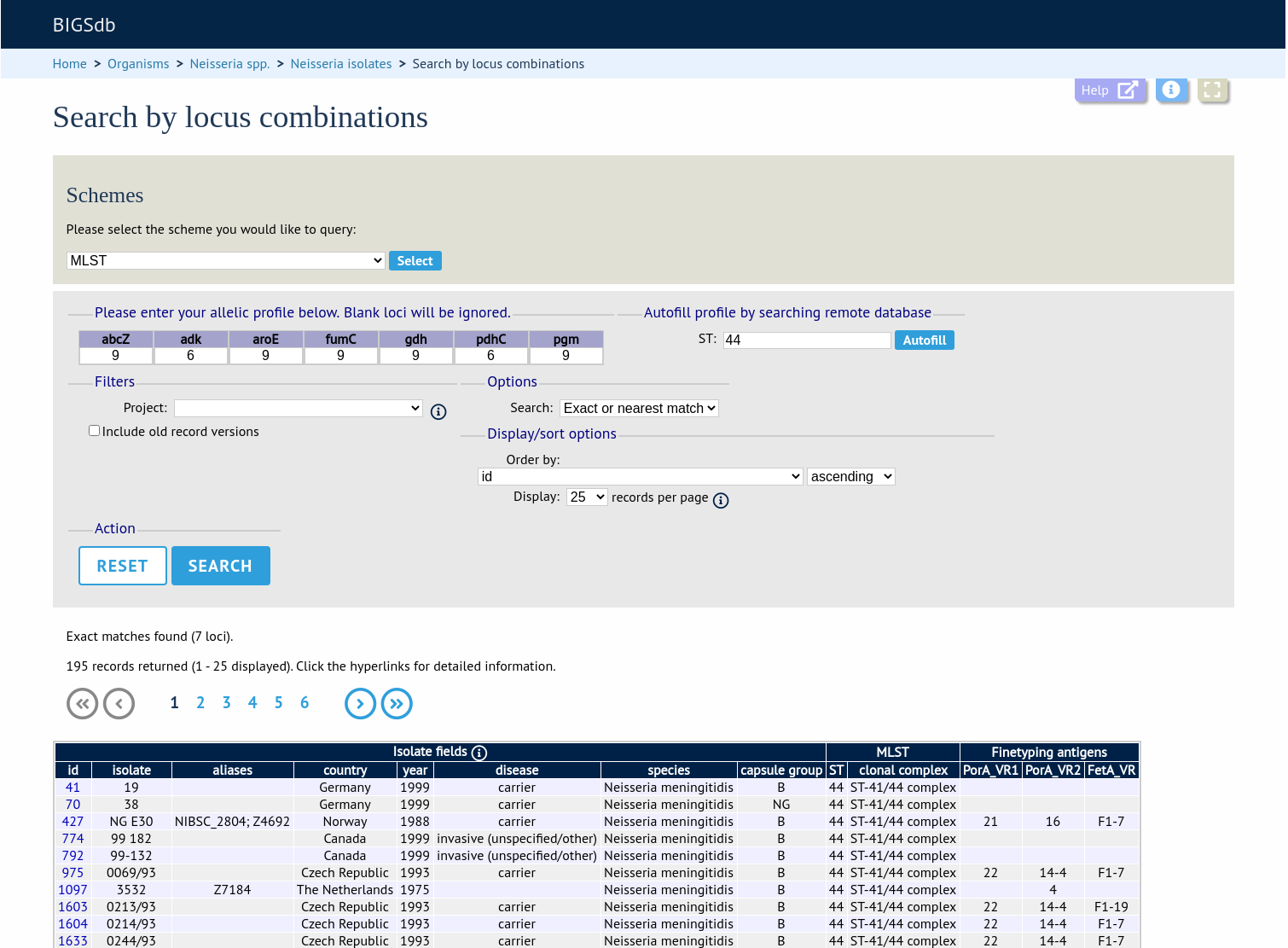
Querying by allelic profile
If a scheme, such as MLST, has been defined for an isolate database it is possible to query the database against complete or partial allelic profiles. Even if no scheme is defined, queries can be made against all loci.
On the index page, click ‘Search by combinations of loci (profiles)’ for any defined scheme. Enter either a partial (any combination of loci) or complete profile.
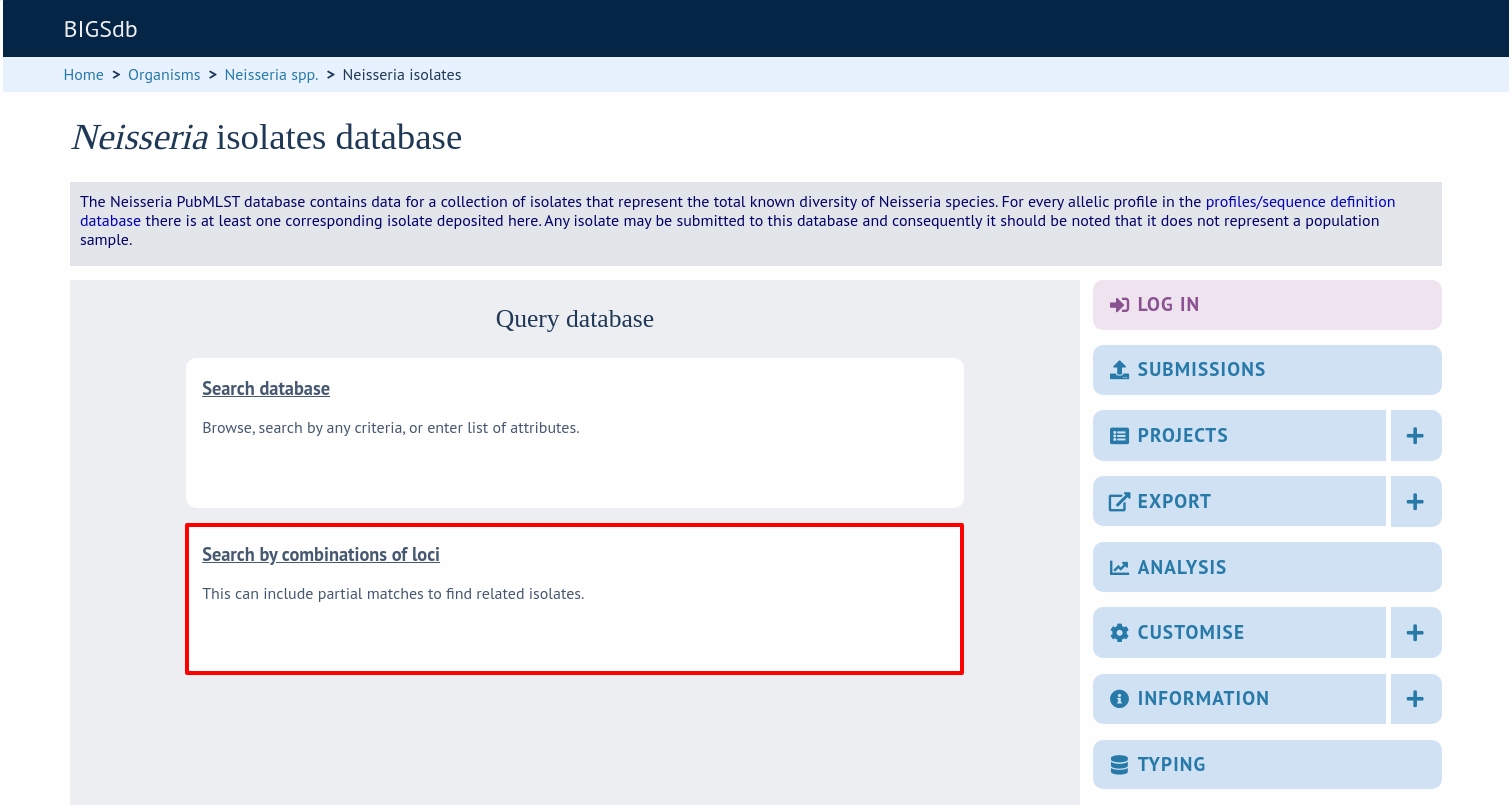
If multiple schemes are defined, you may have to select the scheme you wish to query in the ‘Schemes’ dropdown box and click ‘Select’.
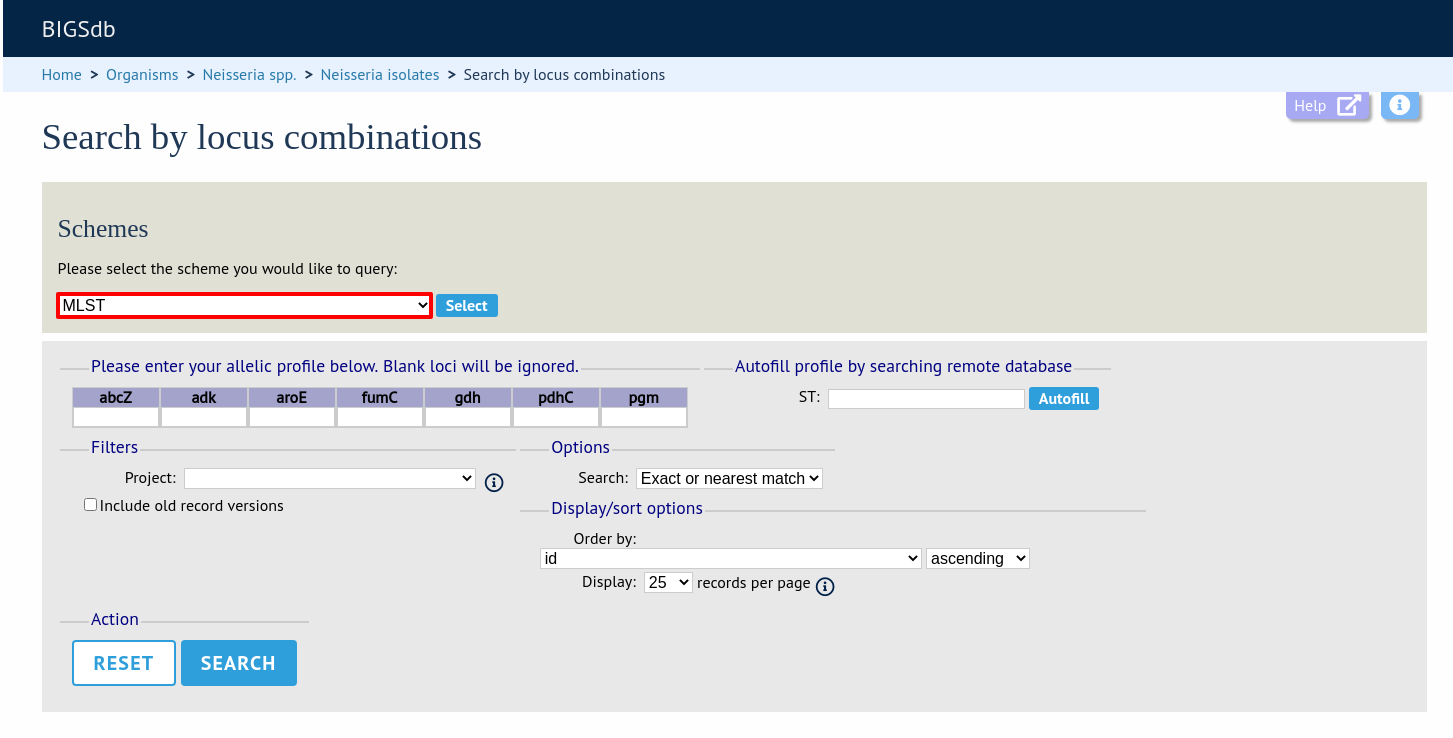
Enter the combination of alleles that you want to query for. Fields can be left blank.
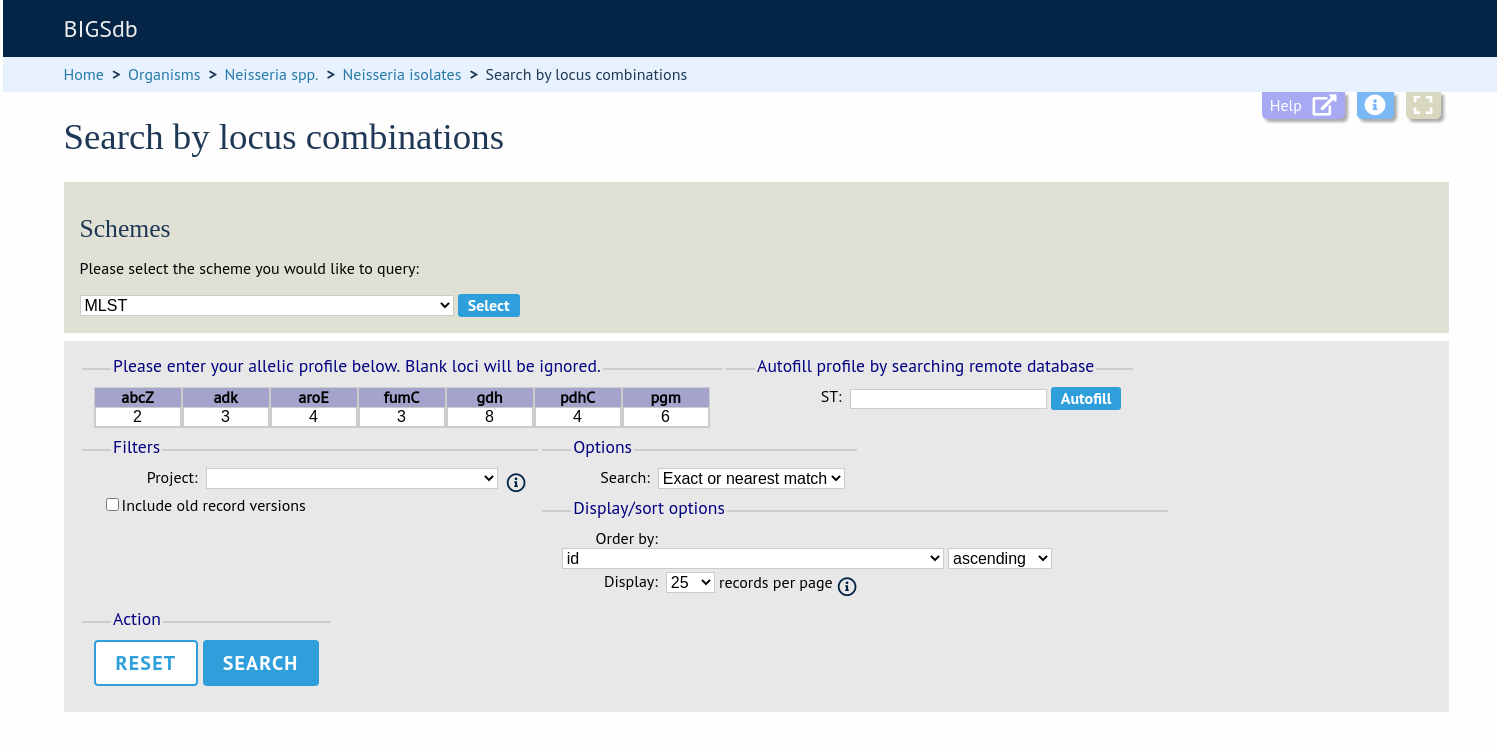
Alternatively, for scheme profiles, you can enter a primary key value (e.g. ST) and select ‘Autofill’ to automatically fill in the associated profile.
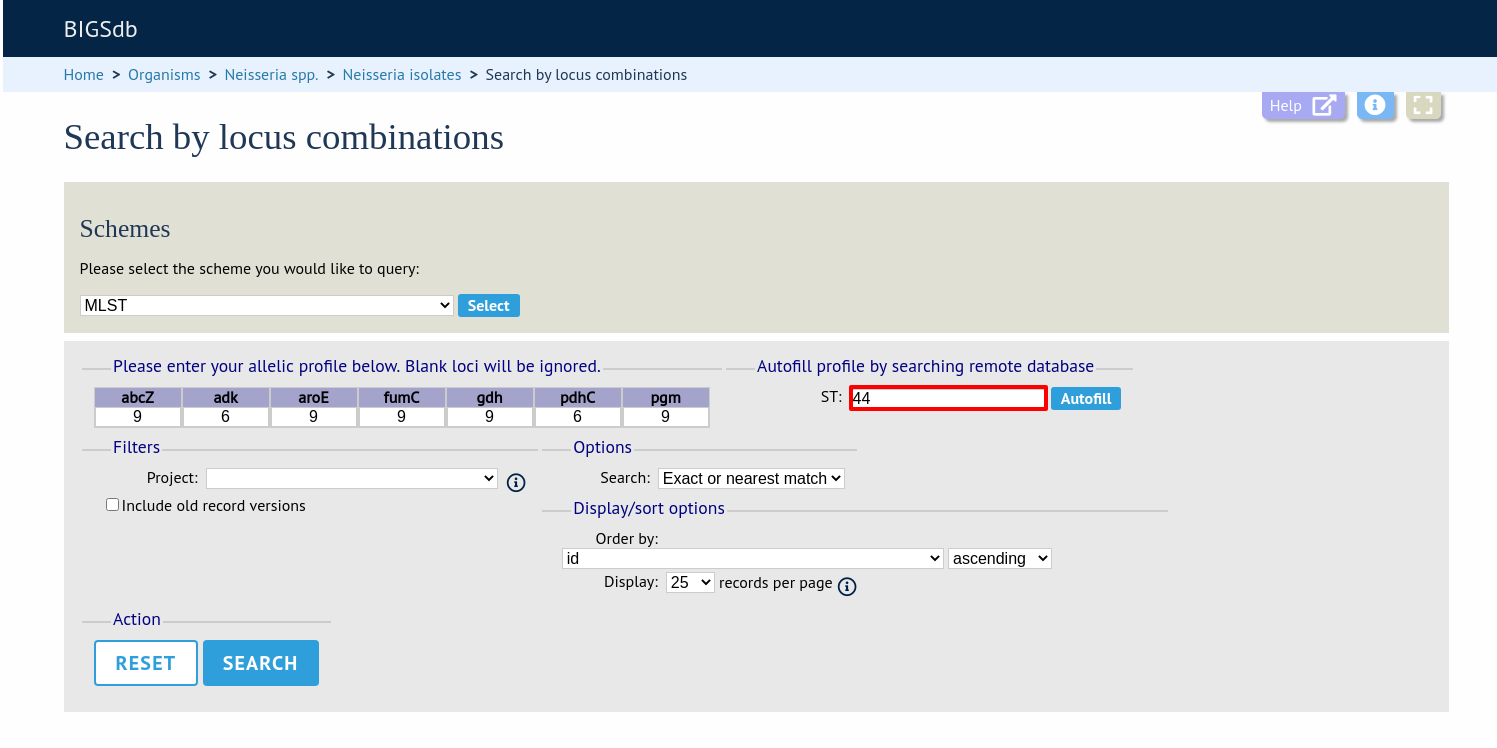
Select the number of loci that you’d like to match in the options dropdown box. Available options are:
Exact or nearest match
Exact match only
x or more matches
y or more matches
z or more matches
Where x,y, and z will range from n-1 to 1 where n is the number of loci in the scheme.
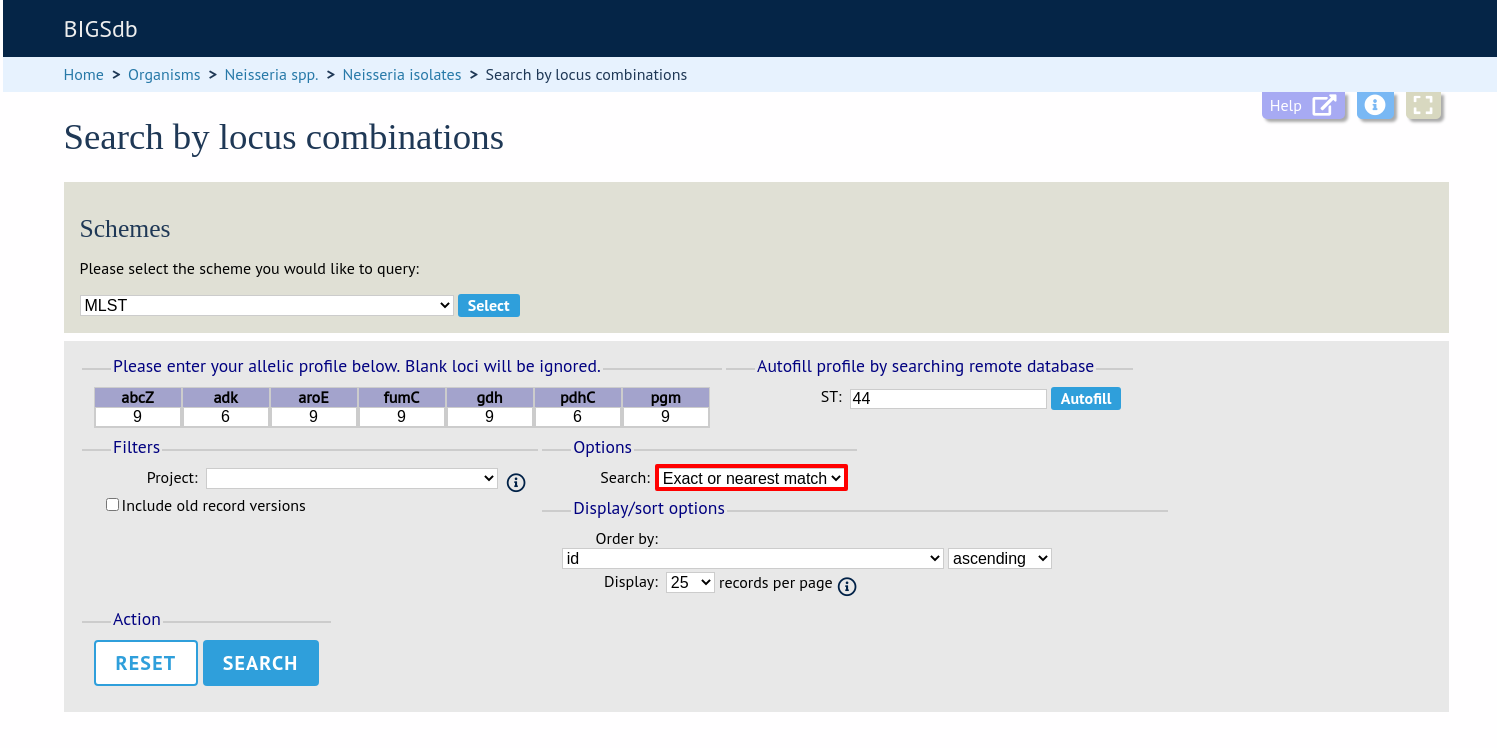
Click ‘Search’.