RESTful Application Programming Interface (API)
The REST API allows third-party applications to retrive data stored within BIGSdb databases or to send new submissions to database curators. To use the REST API, your application will make a HTTP request and parse the response. The response format is JSON (except for routes that request a FASTA or CSV file).
Access to protected resources, i.e. those requiring an account, can be accessed via the API using OAuth authentication.
Passing additional/optional parameters
If you are using a method called with GET, optional parameters can be passed as arguments to the query URL by adding a ‘?’ followed by the first argument and its value (separated by a ‘=’). Additional parameters are separated by a ‘&’, e.g.
https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates?page=2&page_size=100
Methods called with POST require their arguments to be sent as JSON within the post body.
Paging using request headers
Paging of results can be selected using query parameters as described above. In this case, methods that support paging will include a paging object in the JSON response. This will contain links to the next page, last page etc.
The API also supports paging using request headers. The following request headers are supported:
X-OFFSET
X-PER-PAGE
e.g.
curl -i -H "X-PER-PAGE:10" -H "X-OFFSET:0" https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates
If either of these headers are used, the paging object is no longer returned as part of the JSON response. The response will include the following headers:
X-OFFSET
X-PER-PAGE
X-TOTAL-PAGES
Resources
GET / or /db - List site resources
GET /db/{database} - List database resources
GET /db/{database}/classification_schemes - List classification schemes
GET /db/{database}/classification_schemes/{classification_scheme_id} - Retrieve classification scheme information and groups
GET /db/{database}/classification_schemes/{classification_scheme_id}/groups - List groups defined for a classification scheme
GET /db/{database}/classification_schemes/{classification_scheme_id}/groups/{group_id} - List isolates or profiles belonging to a classification scheme group
GET /db/{database}/loci - List loci
GET /db/{database}/loci/{locus} - Retrieve locus record
GET /db/{database}/loci/{locus}/alleles - Retrieve list of alleles defined for a locus
GET /db/{database}/loci/{locus}/alleles_fasta - Download alleles in FASTA format
GET /db/{database}/loci/{locus}/alleles/{allele_id} - Retrieve full allele information
POST /db/{database}/loci/{locus}/sequence - Query sequence to identify allele
POST /db/{database}/sequence - Query sequence to identify allele without specifying locus
GET /db/{database}/sequences - Get summary of defined sequences
GET /db/{database}/schemes - List schemes
GET /db/{database}/schemes/{scheme_id} - Retrieve scheme information
GET /db/{database}/schemes/{scheme_id}/loci - Retrieve scheme loci
GET /db/{database}/schemes/{scheme_id}/fields/{field} - Retrieve information about scheme field
GET /db/{database}/schemes/{scheme_id}/profiles - List allelic profiles defined for scheme
GET /db/{database}/schemes/{scheme_id}/profiles_csv - Download allelic profiles in CSV (tab-delimited) format
GET /db/{database}/schemes/{scheme_id}/profiles/{profile_id} - Retrieve allelic profile record
POST /db/{database}/schemes/{scheme_id}/sequence - Query sequence to extract allele designations/fields for a scheme
POST /db/{database}/schemes/{scheme_id}/designations - Query allelic profile to extract fields for a scheme
GET /db/{database}/isolates - Retrieve list of isolate records
GET /db/{database}/genomes - Retrieve list of isolate records that have genome assemblies
POST /db/{database}/isolates/search - Search isolate database
GET /db/{database}/isolates/{isolate_id} - Retrieve isolate record
GET /db/{database}/isolates/{isolate_id}/allele_designations - Retrieve list of allele designations
GET /db/{database}/isolates/{isolate_id}/allele_designations/{locus} - Retrieve full allele designation record
GET /db/{database}/isolates/{isolate_id}/allele_ids - Retrieve allele identifiers
GET /db/{database}/isolates/{isolate_id}/schemes/{scheme_id}/allele_designations - Retrieve scheme allele designation records
GET /db/{database}/isolates/{isolate_id}/schemes/{scheme_id}/allele_ids - Retrieve list of scheme allele identifiers
GET /db/{database}/isolates/{isolate_id}/contigs - Retrieve list of contigs
GET /db/{database}/isolates/{isolate_id}/contigs_fasta - Download contigs in FASTA format
GET /db/{database}/isolates/{isolate_id}/history - Retrieve isolate update history
GET /db/{database}/contigs/{contig_id} - Retrieve contig record
GET /db/{database}/fields - Retrieve list of isolate provenance field descriptions
GET /db/{database}/fields/{field} - Retrieve values set for a provenance field
GET /db/{database}/users/{user_id} - Retrieve user information
GET /db/{database}/curators - Retrieve list of curators of the database
GET /db/{database}/projects - Retrieve list of projects
GET /db/{database}/projects/{project_id} - Retrieve project information
GET /db/{database}/projects/{project_id}/isolates - Retrieve list of isolates belonging to a project
GET /db/{database}/submissions - Retrieve list of submissions
POST /db/{database}/submissions - Create new submission
GET /db/{database}/submissions/{submission_id} - Retrieve submission record
DELETE /db/{database}/submissions/{submission_id} - Delete submission record
GET /db/{database}/submissions/{submission_id}/messages - Retrieve submission correspondence
POST /db/{database}/submissions/{submission_id}/messages - Add submission correspondence
GET /db/{database}/submissions/{submission_id}/files - retrieve list of supporting files uploaded for submission
POST /db/{database}/submissions/{submission_id}/files - Upload submission supporting file
GET /db/{database}/submissions/{submission_id}/files/{filename} - Download submission supporting file
DELETE /db/{database}/submissions/{submission_id}/files/{filename} - Delete submission supporting file
GET / or /db - List site resources
Required route parameters: None
Optional query parameters: None
Example request URI: https://rest.pubmlst.org/
Response: List of resource groupings (ordered by name). Groups may consist of paired databases for sequence definitions and isolate data, or any set of related resources. Each group contains:
name [string] - short name (usually a single word)
description [string] - fuller description
databases [array] - list of database objects, each consists of three key/value pairs:
name [string] - name of database config
description [string] - short description of resource
href [string] - URI to access resource
GET /db/{database} - List database resources
These will vary depending on whether the resource is an isolate or a sequence definition database.
Required route parameter: database [string] - Database configuration name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates
Response: Object containing a subset of the following key/value pairs:
GET /db/{database}/classification_schemes - List classification schemes
Required route parameter: database [string] - Database configuration name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/classification_schemes
Response: Object containing:
records [integer] - Number of classification schemes.
classification_schemes [array] - List of URIs to classificaton schemes.
GET /db/{database}/classification_schemes/{classification_scheme_id} - Retrieve classification scheme information and groups
Required route parameters:
database [string] - Database configuration name
classification_scheme_id [integer] - Classification scheme id number
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/classification_schemes/1
Response: Object containing some or all of:
id [integer] - Classification scheme id
name [text] - Name of classification scheme
description [text] - Description of classification scheme
relative_threshold [boolean] - True if a relative thresold is used
inclusion_threshold [integer] - The threshold for number of loci difference used to group
groups [string] (sequence definition databases only) - URI to list of groups
id [integer] - group id
profiles [array] - list of URIs to profiles belonging to the group
GET /db/{database}/classification_schemes/{classification_scheme_id}/groups - List groups defined for a classification scheme
Sequence definition databases only.
Required route parameters:
database [string] - Database configuration name
classification_scheme_id [integer] - Classification scheme id number
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/classification_schemes/1/groups
Response: Object containing of:
records [integer] - Number of groups
groups [array] - List of URIs to classification group records.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/classification_schemes/{classification_scheme_id}/groups/{group_id} - List isolates or profiles belonging to a classification scheme group
Required route parameters:
database [string] - Database configuration name
classification_scheme_id [integer] - Classification scheme id number
group_id [integer] - Group id number
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/classification_schemes/4/groups/65
Response: Object containing some of:
records [integer] - Number of isolates or profiles
isolates (isolate database only) [array] - List of URIs to isolate records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
profiles (sequence definition databases only) [array] - List of URIs to profile records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/loci - List loci
Required route parameter: database [string] - Database configuration name
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
alleles_added_after [date] - Include only loci with alleles added after (but not on) specified date (ISO 8601 format). Only recognized in sequence definition databases.
alleles_updated_after [date] - Include only loci with alleles last modified after (but not on) specified date (ISO 8601 format). Only recognized in sequence definition databases.
alleles_added_reldate [integer] - Include only loci with alleles added within the number of days specified. Only recognized in sequence definition databases.
alleles_updated_reldate [integer] - Include only loci with alleles last modified within the number of days specified. Only recognized in sequence definition databases.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/loci
Response: Object containing:
records [integer] - Number of loci
loci [array] - List of URIs to defined locus records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
Note
See also the scheme specific version, allowing filtering by date of last allele update for just the loci that are members of a scheme.
GET /db/{database}/loci/{locus} - Retrieve locus record
Provides information about a locus, including links to allele sequences (in seqdef databases).
Required route parameters:
database [string] - Database configuration name
locus [string] - Locus name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/loci/abcZ
Response: Object containing a subset of the following key/value pairs:
id [string] - locus name
data_type [string] - ‘DNA’ or ‘peptide’
allele_id_format [string] - ‘integer’ or ‘text’
allele_id_regex [string] - regular expression constraining allele ids
common_name [string]
aliases [array] - list of alternative names of the locus
length_varies [boolean]
length [integer] - length if alleles are of a fixed length
coding_sequence [boolean]
orf [integer] - 1-6
schemes [array] - list of scheme objects, each consisting of:
scheme [string] - URI to scheme information
description [string]
min_length [integer] (seqdef databases) - minimum length for variable length loci
max_length [integer] (seqdef databases) - maximum length for variable length loci
alleles [string] (seqdef databases) - URI to list of allele records
alleles_fasta [string] (seqdef databases) - URI to FASTA file of all alleles of locus
curators [array] (seqdef databases) - list of URIs to user records of curators of the locus
publications [array] (seqdef databases) - list of PubMed id numbers of papers describing the locus
full_name [string] (seqdef databases)
product [string] (seqdef databases)
description [string] (seqdef databases)
extended_attributes [array] (seqdef databases) - list of extended attribute objects. Each consists of a subset of the following fields:
field [string] - field name
value_format [string] - ‘integer’, ‘text’, or ‘boolean’
value_regex [string] - regular expression constraining value
description [string] - description of field
length [integer] - maximum length of field
required [boolean]
allowed_values [array] - list of allowed values
genome_position [integer] (isolate databases)
GET /db/{database}/loci/{locus}/alleles - Retrieve list of alleles defined for a locus
Required route parameters:
database [string] - Database configuration name
locus [string] - Locus name
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
include_records [integer] - Set to non-zero value to include array of allele records rather than links.
extended [integer] - Set to non-zero value to include extended attributes if defined (only if include_records is selected).
variation [integer] - Set to non-zero value to include defined single amino-acid variant (SAV) and/or single nucleotide variant (SNP) information if defined for the locus (only if include_records is selected).
added_after [date] - Include only alleles added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Include only alleles added within the specified number of days.
added_on [date] - Include only alleles added on specified date (ISO 8601 format).
updated_after [date] - Include only alleles last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Include only alleles updated within the specified number of days.
updated_on [date] - Include only alleles last modified on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/loci/abcZ/alleles
Response: Object containing:
records [integer] - Number of alleles.
last_updated [date] - Latest allele addition/modification date (ISO 8601 format).
alleles [array] - If include_records = 0 this is a list of URIs to defined allele records. If include_records = 1 then this is a list of allele objects (with values as used in single allele records). Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/loci/{locus}/alleles_fasta - Download alleles in FASTA format
Required route parameters:
database [string] - Database configuration name
locus [string] - Locus name
Optional parameters:
added_after [date] - Include only alleles added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Include only alleles added within the specified number of days.
added_on date [date] - Include only alleles added on specified date (ISO 8601 format).
updated_after [date] - Include only alleles last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Include only alleles last modified within the specified number of days.
updated_on [date] - Include only alleles last modified on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/loci/abcZ/alleles_fasta
Response: FASTA format file of allele sequences
GET /db/{database}/loci/{locus}/alleles/{allele_id} - Retrieve full allele information
Required route parameters:
database [string] - Database configuration name
locus [string] - Locus name
allele_id [string] - Allele identifier
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/loci/abcZ/alleles/5
Response: Object containing the following key/value pairs:
locus [string] - URI to locus description
allele_id [string] - allele identifier
sequence [string] - sequence
status [string] - either ‘Sanger trace checked’, ‘WGS: manual extract’, ‘WGS: automated extract’, or ‘unchecked’
sender [string] - URI to user details of sender
curator [string] - URI to user details of curator
date_entered [string] - record creation date (ISO 8601 format)
datestamp [string] - last updated date (ISO 8601 format)
POST /db/{database}/loci/{locus}/sequence - Query sequence to identify allele
Required route parameters:
database [string] - Database configuration name
locus [string] - Locus name
Required additional parameters (JSON-encoded in POST body):
sequence [string] - Sequence string or base64-encoded FASTA file
Optional parameters (JSON-encoded in POST body):
details [true/false] - Return detailed exact match parameters
base64 [true/false] - Sequence is a base64-encoded FASTA file
Response: Object containing the following key/value pairs:
exact_matches [array] - list of match objects, each consisting of:
allele_id
href - URI to allele record.
additionally if ‘details’ parameter passed:
start - start position on query
end - end position on query
orientation - forward/reverse
length - length of matched allele
contig - contig name if FASTA file is uploaded
If the locus is linked to field data in client isolate databases, there may also be an object called ‘linked_data’ containing values and frequencies of the field for the returned allele.
best_match [object] - consisting of key/value pairs (if no exact matches)
allele_id
href - URI to allele record.
start - start position on query (predicted taking account of allele length)
end - end position on query (predicted taking account of allele length)
orientation - forward/reverse
length - length of matched allele
alignment - length of BLAST alignment
mismatches - number of mismatches
identity - %identity of match
gaps - number of gaps in alignment
POST /db/{database}/sequence - Query sequence to identify allele without specifying locus
Required route parameters:
database [string] - Database configuration name
Required additional parameters (JSON-encoded in POST body):
sequence [string] - Sequence string or base64-encoded FASTA file
Optional parameters (JSON-encoded in POST body):
details [true/false] - Return detailed exact match parameters
base64 [true/false] - Sequence is a base64-encoded FASTA file
Response:
exact_matches [object] consisting of locus keys, each consisting of array of match objects consisting of:
allele_id
href - URI to allele record.
additionally if ‘details’ parameter passed:
start - start position on query
end - end position on query
orientation - forward/reverse
length - length of matched allele
contig - contig name if FASTA file is uploaded
If the locus is linked to field data in client isolate databases, there may also be an object called ‘linked_data’ containing values and frequencies of the field for the returned allele.
Note
This method only supports exact matches. If no match is indicated for a specific locus, use the locus-specific call to identify the closest match.
GET /db/{database}/sequences - Get summary of defined sequences
Required route parameter: database [string] - Database configuration name
Optional parameters:
added_after [date] - Count only alleles added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Count only alleles added within the specified number of days.
added_on [date] - Count only alleles added on specified date (ISO 8601 format).
updated_after [date] - Count only alleles last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Count only alleles last modified within the specified number of days.
updated_on [date] - Count only allele updated on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/sequences
Response: Object containing a subset of the following key/value pairs:
loci [string] - URI to list of loci
records [integer] - Number of alleles defined
last_updated [date] - Latest allele addition/modification date (ISO 8601 format).
GET /db/{database}/schemes - List schemes
Required route parameter: database [string] - Database configuration name
Optional parameters:
with_pk [integer] - Set to non-zero value to only show indexed schemes, i.e. those with a primary key field that defines each unique combination of alleles, e.g. MLST.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes
Response:
records [integer] - Number of schemes
schemes [array] - list of scheme objects, each containing:
scheme [string] - URI to scheme information
description [string]
GET /db/{database}/schemes/{scheme_id} - Retrieve scheme information
Includes links to allelic profiles (in seqdef databases, if appropriate). Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id number
Optional parameters:
added_after [date] - Count only profiles added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Count only profiles added within the specified number of days.
added_on [date] - Count only profiles added on specified date (ISO 8601 format).
updated_after [date] - Count only profiles last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Count only profiles last modified within the specified number of days.
updated_on [date] - Count only profiles updated on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1
Response: Object containing a subset of the following key/value pairs:
id [integer]
description [string]
locus_count [integer] - number of loci belonging to scheme
loci [array] - list of URIs to locus descriptions
has_primary_key_field [boolean]
fields [array] - list of URIs to scheme field descriptions
primary_key_field [string] - URI to primary key field description
profiles [string] - URI to list of profile definitions (only seqdef databases)
profiles_csv [string] - URI to tab-delimited file of all scheme profiles
curators [array] (seqdef databases) - list of URIs to user records of curators of the scheme
records [integer] - Number of profiles
last_added [date] - Latest profile addition/modification date (ISO 8601 format).
last_updated [date] - Latest profile addition/modification date (ISO 8601 format).
GET /db/{database}/schemes/{scheme_id}/loci - Retrieve scheme loci
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id number
Optional parameters:
alleles_added_after [date] - Include only loci with alleles added after (but not on) specified date (ISO 8601 format). Only recognized in sequence definition databases.
alleles_added_reldate [integer] - Include only loci with alleles added within the specified number of days. Only recognized in sequence definition databases.
alleles_updated_after [date] - Include only loci with alleles last modified after specified date (ISO 8601 format). Only recognized in sequence definition databases.
alleles_updated_reldate [integer] - Include only loci with alleles last modified within the specified number of days. Only recognized in sequence definition databases.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/loci
Response: Object containing:
records [integer] - Number of loci
loci [array] - List of URIs to defined locus records.
GET /db/{database}/schemes/{scheme_id}/fields/{field} - Retrieve information about scheme field
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id number
field [string] - Field name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/fields/ST
Response: Object containing the following key/value pairs:
field [string] - field name
type [string] - data type of field (integer or text)
primary_key [boolean] - true if field is the scheme primary key
GET /db/{database}/schemes/{scheme_id}/profiles - List allelic profiles defined for scheme
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
added_after [date] - Include only profiles added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Include only profiles added within the specified number of days.
added_on [date] - Include only profiles added on specified date (ISO 8601 format).
updated_after [date] - Include only profiles last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Include only profiles last modified within the specified number of days.
updated_on [date] - Include only profiles last modified on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/profiles
Response: Object containing:
records [integer] - Number of profiles
last_updated [date] - Latest profile addition/modification date (ISO 8601 format).
profiles [array] - List of URIs to defined profile records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
Note
This method also supports content negotiation. If the request accepts header includes TSV or CSV, then the call is redirected to /db/{database}/schemes/{scheme_id}/profiles_csv.
GET /db/{database}/schemes/{scheme_id}/profiles_csv - Download allelic profiles in CSV (tab-delimited) format
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id
Optional parameters:
added_after [date] - Include only profiles added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Include only profiles added within the specified number of days.
added_on [date] - Include only profiles added on specified date (ISO 8601 format).
updated_after [date] - Include only profiles last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Include only profiles last modified within the specified number of days.
updated_on [date] - Include only profiles last modified on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/profiles_csv
Response: Tab-delimited text file of allelic profiles
GET /db/{database}/schemes/{scheme_id}/profiles/{profile_id} - Retrieve allelic profile record
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id
profile_id [string/integer] - Profile id
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/profiles/11
Response: Object containing the following key/value pairs:
primary_key_term [string/integer] - The field name is the primary key, e.g. ST. The value is the primary key value (primary_id used as an argument).
alleles [object] - list of URIs to allele descriptions
other_scheme_fields [string/integer] - Each scheme field will have its own value if defined. The field name is the name of the field.
sender [string] - URI to user details of sender
curator [string] - URI to user details of curator
date_entered [string] - record creation date (ISO 8601 format)
datestamp [string] - last updated date (ISO 8601 format)
POST /db/{database}/schemes/{scheme_id}/sequence - Query sequence to extract allele designations/fields for a scheme
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id
Required additional parameters (JSON-encoded in POST body):
sequence [string] - Sequence string or base64-encoded FASTA file
Optional parameters (JSON-encoded in POST body):
details [true/false] - Return detailed exact match parameters
partial_matches [true/false] - Return details of partial matches if exact match is not found
base64 [true/false] - Sequence is a base64-encoded FASTA file
Response: Object containing the following key/value pairs:
exact_matches [array] - list of match objects, each consisting of:
allele_id
href - URI to allele record.
additionally if ‘details’ parameter passed:
start - start position on query
end - end position on query
orientation - forward/reverse
length - length of matched allele
contig - contig name if FASTA file is uploaded
If the locus is linked to field data in client isolate databases, there may also be an object called ‘linked_data’ containing values and frequencies of the field for the returned allele.
Example curl call to upload a FASTA file ‘contigs.fasta’ and extract MLST results from Neisseria database:
(echo -n '{"base64":true,"sequence": "'; base64 contigs.fasta; echo '"}') |
curl -s -H "Content-Type: application/json" -X POST "https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/sequence" -d @-
Note
This method only supports exact matches. If no match is indicated for a specific locus, use the locus-specific call to identify the closest match.
POST /db/{database}/schemes/{scheme_id}/designations - Query allelic profile to extract fields for a scheme
Required route parameters:
database [string] - Database configuration name
scheme_id [integer] - Scheme id
Required additional parameters (JSON-encoded in POST body):
designations [object] - consisting of
locus objects each containing an array of alleles (see example)
Response: Object containing the following key/value pairs:
exact_matches [object] - consisting of locus values, each consisting of an array of allele values:
allele_id [string]
If the locus is linked to field data in client isolate databases, there may also be an object called ‘linked_data’ containing values and frequencies of the field for the returned allele.
fields [object] - consisting of key/value pairs of scheme fields (if defined)
Example curl call to query an allelic profile and extract MLST results from Neisseria database:
curl -s -H "Content-Type: application/json" -X POST "https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/schemes/1/designations" -d '{"designations":{"abcZ":[{"allele":"2"}],"adk":[{"allele":"3"}],"aroE":[{"allele":"4"}],"fumC":[{"allele":"3"}],"gdh":[{"allele":"8"}],"pdhC":[{"allele":"4"}],"pgm":[{"allele":"6"}]}}'
GET /db/{database}/isolates - Retrieve list of isolate records
Required route parameter: database [string] - Database configuration name
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
added_after [date] - Include only isolates added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Include only isolates added within the specified number of days.
added_on [date] - Include only isolates added on specified date (ISO 8601 format).
include_old_versions [integer] - Set to 1 to include old record versions (the default is to only include new versions)
updated_after [date] - Include only isolates last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Include only isolates last modified within the specified number of days.
updated_on [date] - Include only isolates updated on specified date (ISO 8601 format).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates
Response: Object containing:
records [integer] - Number of isolates
isolates [array] - List of URIs to isolate records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/isolates/{isolate_id} - Retrieve isolate record
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
Optional parameter:
provenance_only [integer] - Set to non-zero value to only return provenance metadata
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1
Response: Object containing some or all of the following key/value pairs:
provenance [object] - set of key/value pairs. Keys are defined by calling the /fields route route. The fields will vary by database but will always contain the following:
id [integer]
sender [string] - URI to user details of sender
curator [string] - URI to user details of curator
date_entered [string] - record creation date (ISO 8601 format)
datestamp [string] - last updated date (ISO 8601 format)
aliases [array] - list of alternative names for isolate
publications [array] (seqdef databases) - list of PubMed id numbers of papers that refer to the isolate
sequence_bin [object] - consists of the following key/value pairs:
contigs_fasta [string] - URI to FASTA file containing all the contigs belonging to this isolate
contigs [string] - URI to list of contig records
contig_count [integer] - number of contigs
total_length [integer] - total length of contigs
allele_designations [object] - consists of the following key/value pairs:
allele_ids - URI to list of all allele_id values defined for the isolate
designation_count - number of allele designations defined for the isolate
full_designations - URI to list of full allele designation records
schemes [array] - list of scheme objects, each containing some of the following:
description [string] - description of scheme
loci_designated_count [integer] - number of loci within scheme that have an allele designated for this isolate.
allele_ids [string] - URI to list of all allele_id values defined for this scheme for this isolate
full_designations [string] - URI to list of full allele designation records for this isolate
fields [object] - consisting of key/value pairs where the key is the name of each scheme field
classification_schemes [object] - consisting of key/value pairs, where each key is the name of the classification scheme and the value is an object consisting of:
href [string] - URI to classification scheme description
groups [array] - list of group objects consisting of:
group [integer] - group id
records [integer] - number of isolates in group
isolates [string] - URI to classification group record containing URIs to member isolate records
projects [array] - list of project objects, each containing the following:
id [string] - URI to project information
description [string] - description of project
history [string] - URI to isolate history record
new_version [string] - URI to newer version of record
old_version [string] - URI to older version of record
GET /db/{database}/isolates/{isolate_id}/allele_designations - Retrieve list of allele designation records
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/allele_designations
Response: Object containing:
records [integer] - Number of allele designations
allele_designations [array] - List of URIs to allele designation records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/isolates/{isolate_id}/allele_designations/{locus} - Retrieve full allele designation record
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
locus [string] - Locus name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/allele_designations/BACT000065
Response: List of allele_designation objects (there may be multiple designations for the same locus), each containing:
locus [string] - URI to locus description
allele_id [string]
method [string] - either ‘manual’ or ‘automatic’
status [string] - either ‘confirmed’ or ‘provisional’
comments [string]
sender [string] - URI to user details of sender
curator [string] - URI to user details of curator
datestamp [string] - last updated date (ISO 8601 format)
GET /db/{database}/isolates/{isolate_id}/allele_ids - Retrieve allele identifiers
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/allele_ids
Response: Object containing:
records [integer] - Number of allele id objects
allele_ids [array] - List of allele id objects, each consisting of a key/value pair where the key is the locus name. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/isolates/{isolate_id}/schemes/{scheme_id}/allele_designations - Retrieve scheme allele designation records
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
scheme_id [integer] - Scheme identifier
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/schemes/1/allele_designations
Response:
records [integer] - Number of allele designation objects
allele_designations [array] - List of allele designation objects for each locus in the specified scheme that has been designated.
GET /db/{database}/isolates/{isolate_id}/schemes/{scheme_id}/allele_ids - Retrieve list of scheme allele identifiers
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
scheme_id [integer] - Scheme identifier
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/schemes/1/allele_ids
Response:
records [integer] - Number of allele id objects
allele_ids [array] - List containing allele id objects for each locus in the specified scheme that has been designated. Each allele_id object contains a key which is the name of the locus with a value that may be either a string, integer or array of strings or integers (required where there are multiple designations for a locus). The data type depends on the allele_id_format set for the specific locus.
GET /db/{database}/isolates/{isolate_id}/contigs - Retrieve list of contigs
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/contigs
Response: Object containing:
records [integer] - Number of contigs
contigs [array] - List of URIs to contig records Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/isolates/{isolate_id}/contigs_fasta - Download contigs in FASTA format
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
Optional parameter:
header [string] - either ‘original_designation’ or ‘id’ (default is ‘id’). This selects whether the FASTA header lines contain the originally uploaded FASTA headers or the sequence bin id numbers.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/contigs_fasta?header=original_designation
Response: FASTA format file of isolate contig sequences
GET /db/{database}/isolates/{isolate_id}/history - Retrieve isolate update history
Required route parameters:
database [string] - Database configuration name
isolate_id [integer] - Isolate identifier
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/1/history
Response: Object containing:
records [integer] - Number of updayes
contigs [array] - List of update objects each consisting of the following key/value pairs:
curator [string] - URI to user details of curator
timestamp [string] - Time of update
actions [array] - List of update descriptions [strings]
GET /db/{database}/genomes - Retrieve list of isolate records that have genome assemblies
Required route parameter: database [string] - Database configuration name
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
added_after [date] - Include only isolates added after (but not on) specified date (ISO 8601 format).
added_reldate [integer] - Include only isolates added within the specified number of days.
added_on [date] - Include only isolates added on specified date (ISO 8601 format).
include_old_versions [integer] - Set to 1 to include old record versions (the default is to only include new versions)
updated_after [date] - Include only isolates last modified after (but not on) specified date (ISO 8601 format).
updated_reldate [integer] - Include only isolates last modified within the specified number of days.
updated_on [date] - Include only isolates updated on specified date (ISO 8601 format).
genome_size [integer] - Filter to only include records with a sequence bin of at least the specified size (default is 500,000bp).
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/genomes
Response: Object containing:
records [integer] - Number of isolates
isolates [array] - List of URIs to isolate records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
POST /db/{database}/isolates/search - Search isolate database
Required route parameters:
database [string] - Database configuration name
Optional parameters (appended to URI):
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Query parameters (JSON-encoded in POST body):
You must include at least one query parameter.
Parameter names in the following forms are supported:
field.{field} - key/value pairs for provenance fields. Supported field names can be found by calling the /fields route. The fields will vary by database.
locus.{locus} - key/value pairs of locus and its allele designation. Supported locus names can be found by calling the /loci route.
scheme.{scheme_id}.{scheme_field} - key/value pairs of scheme fields and their values. Supported field names can be determined by following routes from the /schemes route.
Example method call using curl: The following searches for Neisseria ST-11 isolates from Europe in 2015 (MLST is scheme#1 in this database).
curl -s -H "Content-Type: application/json" -X POST \
"https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/isolates/search" \
-d '{"field.continent":"europe","field.year":2015,"scheme.1.ST":11}'
Response: Object containing:
records [integer] - Number of isolates
isolates [array] - List of URIs to isolate records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/contigs/{contig_id} - Retrieve contig record
Required route parameters:
database [string] - Database configuration name
contig_id [integer] - Contig identifier
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/contigs/180062
Response: Contig object consisting of the following key/value pairs:
id [integer] - contig identifier
isolate_id [integer] - isolate identifier
sequence [string] - contig sequence
length [integer] - length of contig sequence
method [string] - sequencing method
sender [string] - URI to user details of sender
curator [string] - URI to user details of curator
date_entered [string] - record creation date (ISO 8601 format)
datestamp [string] - last updated date (ISO 8601 format)
loci [array] - list of sequence tag objects consisting of:
locus [string] - URI to locus description
locus_name [string]
start [integer]
end [integer]
direction [string] - forward/reverse
complete [boolean] - true/false
GET /db/{database}/fields - Retrieve list of isolate provenance field descriptions
Required route parameters:
database [string] - Database configuration name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/fields
Response: Array of field objects, each consisting of some or all of the following key/value pairs:
name [string] - name of field
type [string] - data type (int, text, date, float)
length [integer] - maximum length of field
required [boolean] - true if field value is required
min [integer] - minimum value for integer values
max [integer] - maximum value for integer values
regex [string] - regular expression that constrains the allowed value of the field
comments [string]
allowed values [array] - list of allowed values for the field
values [string] - URI to list of used field values
GET /db/{database}/fields/{field} - Retrieve values set for a provenance field
Required route parameters:
database [string] - Database configuration name
field [string] - Provenance metadata field name
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/fields/country
Response: Object containing:
records [integer] - Number of values
values [array] - List of values used in isolate records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/users/{user_id} - Retrieve user information
Users may be data submitters or curators.
Required route parameters:
database [string] - Database configuration name
user_id [integer] - User id number
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/users/2
Response: Object containing the following key/value pairs:
id [integer] - user id number
first_name [string]
surname [string]
affiliation [string] - institutional affiliation
email [string] - E-mail address (may be hidden depending on server configuration)
GET /db/{database}/curators - Retrieve list of curators
Required route parameters:
database [string] - Database configuration name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/curators
Response: Object containing:
records [integer] - Number of curators
curators [array] - List of URIs to user records.
GET /db/{database}/projects - Retrieve list of projects
Required route parameter: database [string] - Database configuration name
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/projects
Response:
projects [array] - List of project objects, each containing:
project [string] - URI to project information
description [string]
isolate_count [integer] - number of isolates in project
GET /db/{database}/projects/{project_id} - Retrieve project information
Required route parameters:
database [string] - Database configuration name
project_id [integer] - Project id number
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/projects/3
Response: Object containing a subset of the following key/value pairs:
id [integer]
description [string]
isolates [string] - URI to list of URIs of member isolate records.
GET /db/{database}/projects/{project_id}/isolates - Retrieve list of isolates belonging to a project
Required route parameter:
database [string] - Database configuration name
project_id [integer] - Project id number
Optional parameters:
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/projects/3/isolates
Response: Object containing:
records [integer] - Number of isolates in the project
isolates [array] - List of URIs to isolate records. Pages are 100 records by default. Page size can be modified using the page_size parameter.
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
GET /db/{database}/submissions - retrieve list of submissions
Required route parameter: database [string] - Database configuration name
Optional parameters:
type [string] - either ‘alleles’, ‘profiles’ or ‘isolates’
status [string] - either ‘closed’ or ‘pending’
page [integer]
page_size [integer]
return_all [integer] - Set to non-zero value to disable paging.
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_isolates/submissions
Response: Object containing:
records [integer] - Number of submissions
submissions [array] - List of URIs to submission records
paging [object] - Some or all of the following:
previous - URI to previous page of results
next - URI to next page of results
first - URI to first page of results
last - URI to last page of results
return_all - URI to page containing all results (paging disabled)
POST /db/{database}/submissions - create new submission
Required route parameter: database [string] - Database configuration name
Required additional parameters (JSON-encoded in POST body):
type [string] - either:
alleles (sequence definition databases only)
profiles (sequence definition databases only)
isolates (isolate databases only)
genomes (isolate databases only)
The following are required with the specified database type:
Allele submissions
locus [string] - name of locus
technology [string] - name of sequencing technology: either ‘454’, ‘Illumina’, ‘Ion Torrent’, ‘PacBio’, ‘Oxford Nanopore’, ‘Sanger’, ‘Solexa’, ‘SOLiD’, or ‘other’
read_length [string] - read length of sequencing: either ‘<100’, ‘100-199’, ‘200-299’, ‘300-499’, ‘>500’, or any positive integer (only required for Illumina)
coverage [string] - mean coverage of sequencing: either ‘<20x’, ‘20-49x’, ‘50-99x’, ‘>100x’, or any positive integer (only required for Illumina)
assembly [string] - assembly method: either ‘de novo’ or ‘mapped’
software [string] - name of assembly software
sequences [string] - either single raw sequence or multiple sequences in FASTA format
Profile submissions
scheme_id [integer] - scheme id number
profiles [string] - tab-delimited profile data - this should include a header line containing the name of each locus
Isolate submissions
isolates [string] - tab-delimited isolate data - this should include a header line containing each field or locus included
Genome submissions
isolates [string] - tab-delimited isolate data - this should include a header line containing each field or locus included as well as for ‘assembly_filename’ and ‘sequence_method’. The ‘sequence_method’ should be either ‘454’, ‘Illumina’, ‘Ion Torrent’, ‘PacBio’, ‘Oxford Nanopore’, ‘Sanger’, ‘Solexa’, ‘SOLiD’, or ‘other’. Following submission, contig files should be uploaded with the same names as set for ‘assembly_filename’. This can be done using the file upload route.
Optional parameters:
message [string] - correspondence to the curator
email [integer] - set to 1 to enable E-mail updates (E-mails will be sent to the registered user account address).
Response: Object containing:
submission - URI to submission record
For genome submissions, the response object will also contain:
missing_files [array] - List of filenames that need to be uploaded to complete the submission. These filenames are defined in the ‘assembly_filename’ field of the isolate record upload. The files should contain the contig assemblies.
message [string] - ‘Please upload missing contig files to complete submission.’
GET /db/{database}/submissions/{submission_id} - Retrieve submission record
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/submissions/BIGSdb_20151013081836_14559_14740
Response: Object containing some of the following:
id [string] - Submission id
type [string] - Either ‘alleles’, ‘profiles’, ‘isolates’
date_submitted [string] - Submission date (ISO 8601 format)
datestamp [string] - Last updated date (ISO 8601 format)
submitter [string] - URI to user details of submitter
curator [string] - URI to user details of curator
status [string] - either ‘started’, ‘pending’, or ‘closed’
outcome [string] - either ‘good’ (data uploaded), ‘bad’ (data rejected), or ‘mixed’ (parts of submission accepted)
correspondence [array] - List of correspondence objects in time order. Each contains:
user [string] URI to user details of user
timestamp [string]
message [string]
Allele submissions
locus [string] - name of locus
technology [string] - name of sequencing technology: either ‘454’, ‘Illumina’, ‘Ion Torrent’, ‘PacBio’, ‘Oxford Nanopore’, ‘Sanger’, ‘Solexa’, ‘SOLiD’, or ‘other’
read_length [string] - read length of sequencing: either ‘<100’, ‘100-199’, ‘200-299’, ‘300-499’, ‘>500’, or any positive integer (only required for Illumina)
coverage [string] - mean coverage of sequencing: either ‘<20x’, ‘20-49x’, ‘50-99x’, ‘>100x’, or any positive integer (only required for Illumina)
assembly [string] - assembly method: either ‘de novo’ or ‘mapped’
software [string] - name of assembly software
seqs [array] - List of sequence objects each containing:
seq_id [string] - Sequence identifier
assigned_id [string] - Allele identifier if uploaded to the database (otherwise undefined)
status [string] - Either ‘pending’, ‘assigned’, or ‘rejected’
sequence [string]
Profile submissions
scheme [string] - URI to scheme information
profiles [array] - List of profile record objects. Each contains:
profile_id [string] - Record identifier
assigned_id [string] - Profile identifier if uploaded to the database (otherwise undefined)
status [string] - Either ‘pending’, ‘assigned’, or ‘rejected’
designations [object] containing key/value pairs for each locus containing the allele identifier
Isolate submissions
isolates [array] - List of isolate record objects. Each contains key/value pairs for included fields.
DELETE /db/{database}/submissions/{submission_id} - Delete submission record
You must be the owner and the record must be closed.
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/submissions/BIGSdb_20151013081836_14559_14740
Response: message [string] - ‘Submission deleted.’
GET /db/{database}/submissions/{submission_id}/messages - Retrieve submission correspondence
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/submissions/BIGSdb_20151013081836_14559_14740/messages
Response: Array of correspondence objects in time order. Each contains:
user [string] URI to user details of user
timestamp [string]
message [string]
POST /db/{database}/submissions/{submission_id}/messages - Add submission correspondence
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
Required additional parameter (JSON-encoded in POST body):
message [string] - Message text
Optional parameters: None
Response: message [string] - ‘Message added.’
GET /db/{database}/submissions/{submission_id}/files - Retrieve list of supporting files uploaded for submission
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
Optional parameters: None
Example request URI: https://rest.pubmlst.org/db/pubmlst_neisseria_seqdef/submissions/BIGSdb_20151013081836_14559_14740/files
Response: Array of URIs to files
POST /db/{database}/submissions/{submission_id}/files - Upload submission supporting file
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
Required additional parameters (JSON-encoded in POST body):
filename [string] - Name of file to store within submission
upload [base64 encoded data] - Raw file data
Optional parameters: None
Response: message [string] - ‘File uploaded.’
GET /db/{database}/submissions/{submission_id}/files/{filename} - Download submission supporting file
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
filename [string] - Name of file
Optional parameters: None
Response: File download
DELETE /db/{database}/submissions/{submission_id}/files/{filename} - Delete submission supporting file
Required route parameters:
database [string] - Database configuration name
submission_id [string] - Submission id
filename [string] - Name of file
Optional parameters: None
Response: message [string] - ‘File deleted.’
Authentication
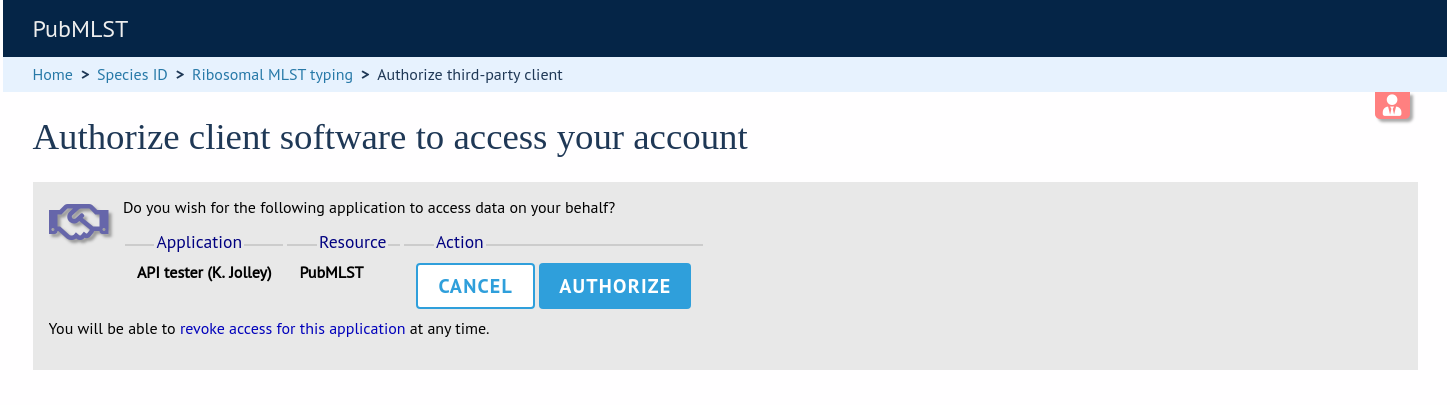
Protected resources, i.e. those requiring a user to log in, can be accessed via the API using OAuth (1.0A) authentication (see IETF RFC5849 for details). Third-party client software has to be registered with the BIGSdb site before they can access authenticated resources. The overall three-legged flow works as follows:
Developer signs up and gets a consumer key and consumer secret specific to their application.
Application gets a request token and directs user to authorization page on BIGSdb.
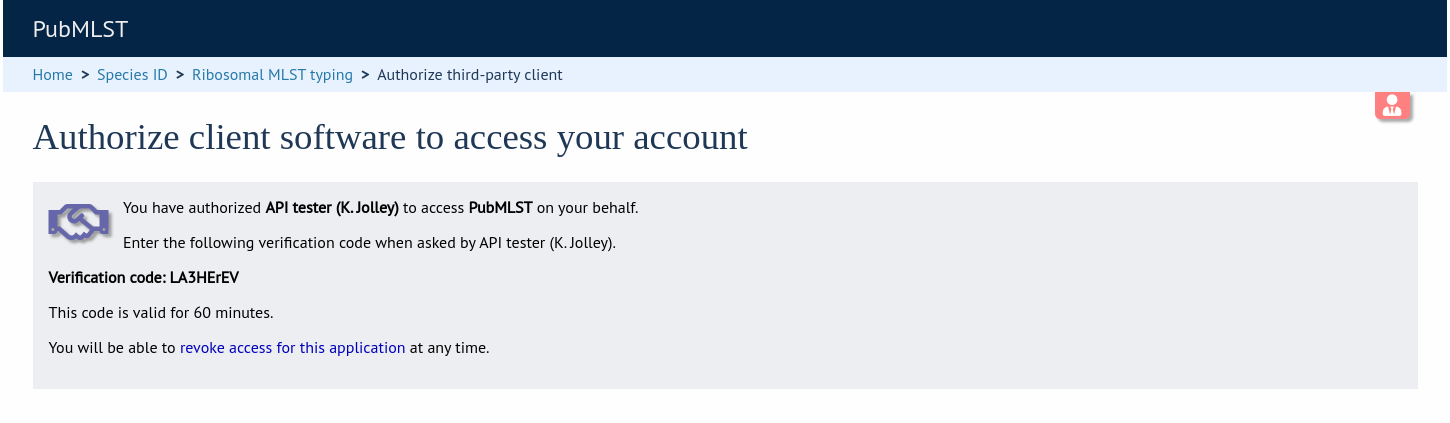
BIGSdb asks user for authorization for application to access specific resource using their credentials. A verifier code is provided.
The application exchanges the request token and OAuth verifier code for an access token and secret (these do not expire but may be revoked by the user or site admin).
Application uses access token/secret to request session token (this is valid for 12 hours).
All calls to access protected resources are signed using the session token/secret and consumer key/secret.
It is recommended that application developers use an OAuth library to generate and sign requests.
Note
There are Python and Perl example scripts available at https://github.com/kjolley/BIGSdb/tree/develop/scripts/rest_examples to demonstrate and test OAuth authentication.
Developer sign up to get a consumer key
Application developers should apply to the site administrator of the site running BIGSdb. The administrator can generate a key and secret using a script - both of these will need to be used by the application to sign requests.
The client id is usually a 24 character alphanumeric string. The secret is usually a 42 character alphanumeric (including punctuation) string, e.g.
client_id: efKXmqp2D0EBlMBkZaGC2lPf
client_secret: F$M+fQ2AFFB2YBDfF9fpHF^qSWJdmmN%L4Fxf5Gur3
Getting a request token
Relative URL: /db/{database}/oauth/get_request_token
Supported method: GET
The application uses the consumer key to obtain a request token. The request token is a temporary token used to initiate user authorization for the application and will expire in 60 minutes. The request needs to contain the following parameters and to be signed using the consumer secret:
oauth_consumer_key
oauth_request_method (‘GET’)
oauth_request_url (request URL)
oauth_signature_method (‘HMAC-SHA1’)
oauth_signature
oauth_timestamp (UNIX timestamp - seconds since Jan 1 1970) - this must be within 600 seconds of the current time.
oauth_callback (‘oob’ for desktop applications)
oauth_nonce (random string)
oauth_version (‘1.0’)
If the application has been registered and has been granted permission to access the specific resource, a JSON response will be returned containing the following parameters:
- oauth_token
This is the request token. It is usually a 32 character alphanumeric string.
e.g. fKFm0WNhCfbEX8zQm6qhDA8K23FOWDGE
- oauth_token_secret
This is the secret associated with the request token. It is usually a 32 character alphanumeric string.
e.g. aZ0fncP7i5w5jlebdK5zyQ4vrRRVcdnv
- oauth_callback_confirmed
This parameter is always set to true.
Getting an access token
Relative URL: /db/{database}/oauth/get_access_token
Supported method: GET
The application uses the request token, verifier code and its consumer key to obtain an access token. The access token does not expire but can be revoked by either the end user or the site administrator. The request needs to contain the following parameters and to be signed using the consumer secret and request token secret:
oauth_consumer_key
oauth_request_method (‘GET’)
oauth_request_url (request URL)
oauth_signature_method (‘HMAC-SHA1’)
oauth_signature
oauth_token (request token)
oauth_timestamp (UNIX timestamp - seconds since Jan 1 1970) - this must be within 600 seconds of the current time.
oauth_nonce (random string)
oauth_version (‘1.0’)
If the application has been registered and has been granted permission to access the specific resource, a JSON response will be returned containing the following parameters:
- oauth_token
This is the access token. It is usually a 32 character alphanumeric string.
e.g. SDrC74ZVl5SYSqY8lWZqrRxnyDnNGVFO
- oauth_token_secret
This is the secret associated with the access token. It is usually a 32 character alphanumeric string.
e.g. tYI2SPzgiO02IRVzW4JR1ez6Vvm4gVyv
Getting a session token
Relative URL: /db/{database}/oauth/get_session_token
Supported method: GET
The application uses the access token and its consumer key to obtain a session token. The session token is valid for 12 hours before it expires. The request needs to contain the following parameters and to be signed using the consumer secret and access token secret:
oauth_consumer_key
oauth_request_method (‘GET’)
oauth_request_url (request URL)
oauth_signature_method (‘HMAC-SHA1’)
oauth_signature
oauth_token (access token)
oauth_timestamp (UNIX timestamp - seconds since Jan 1 1970) - this must be within 600 seconds of the current time.
oauth_nonce (random string)
oauth_version (‘1.0’)
If the application has been registered and has been granted permission to access the specific resource, a JSON response will be returned containing the following parameters:
- oauth_token
This is the session token. It is usually a 32 character alphanumeric string.
e.g. H8CjIS8Ikq6hwCUqUfF1l4pTaCYl8Ljw
- oauth_token_secret
This is the secret associated with the session token. It is usually a 32 character alphanumeric string.
e.g. RfponbaNPO7tkZ2miHFISk0pMndePNfJ
Accessing protected resources
The application uses the session token and its consumer key to access a protected resource. The request needs to contain the following parameters and to be signed using the consumer secret and session token secret:
oauth_consumer_key
oauth_request_method (‘GET’)
oauth_request_url (request URL)
oauth_signature_method (‘HMAC-SHA1’)
oauth_signature
oauth_token (session token)
oauth_timestamp (UNIX timestamp - seconds since Jan 1 1970) - this must be within 600 seconds of the current time.
oauth_nonce (random string)
oauth_version (‘1.0’)