PhyloViz
PhyloViz Online is a tool for generating and visualising minimum-spanning trees. Datasets can include metadata which allows nodes in the resultant tree to be coloured interactively.
PhyloViz can be accessed by selecting the ‘Analysis’ section on the main contents page.
Jump to the ‘Third party’ category, follow the link to PhyloViz, then click ‘Launch PhyloViz’.

Alternatively, it can be accessed following a query by clicking the ‘PhyloViz’ button at the bottom of the results table. Isolates returned from the query will be automatically selected within the PhyloViz interface.
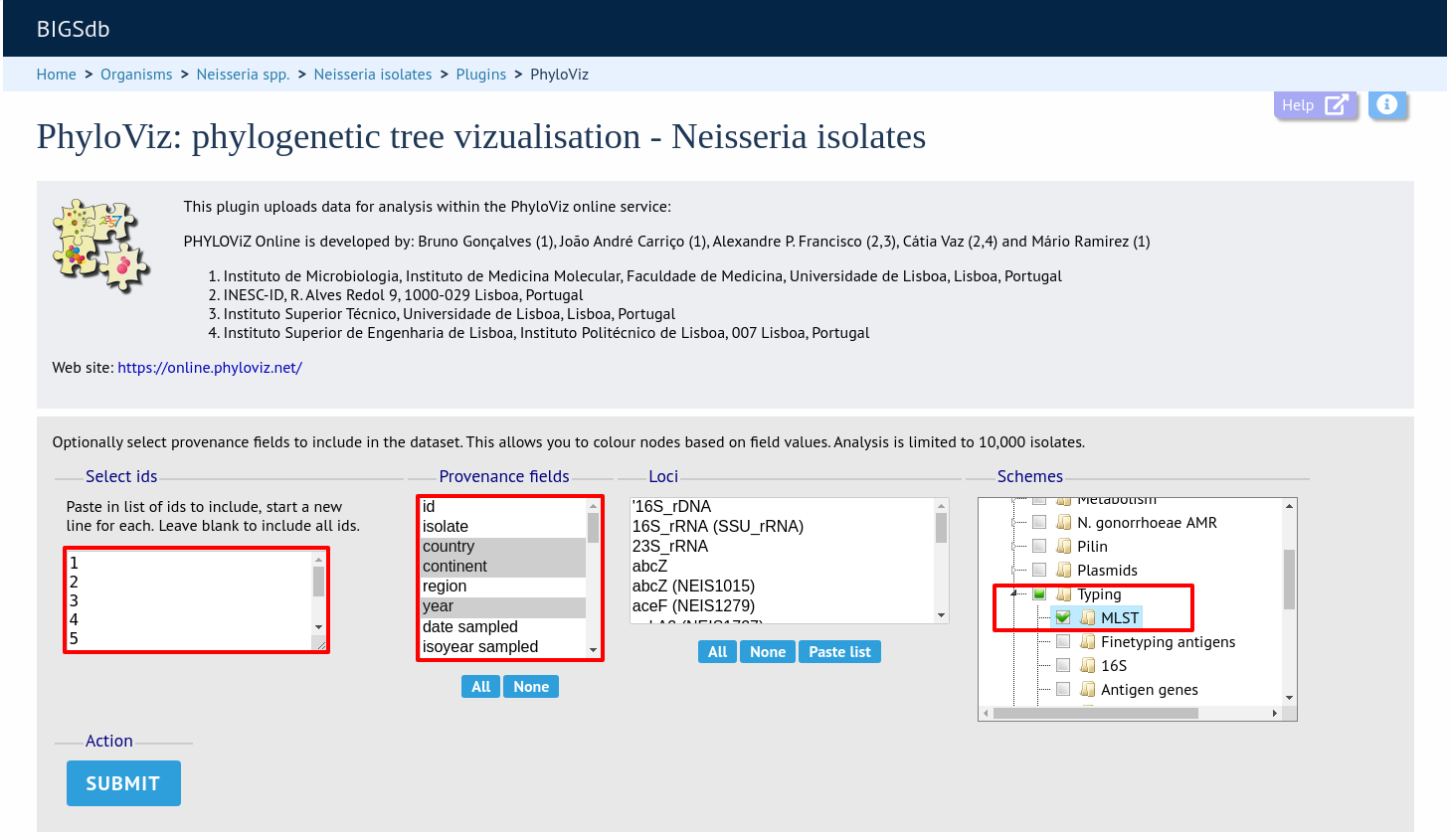
Select the isolates to include. The tree can be generated from allelic profiles of any selection of loci, or more conveniently, you can select a scheme in the scheme selector to include all loci belong to that scheme.
Provenance fields can be selected to be included as metadata for use in colouring nodes - select any fields you wish to include. Multiple selections can be made by holding down Shift or Ctrl while selecting. Click ‘Submit’ to start the analysis.
The necessary files will be generated immediately. When this has finished, click the button launch ‘Launch PhyloViz’.

The tree will be sent to and rendered within the PhyloViz website.
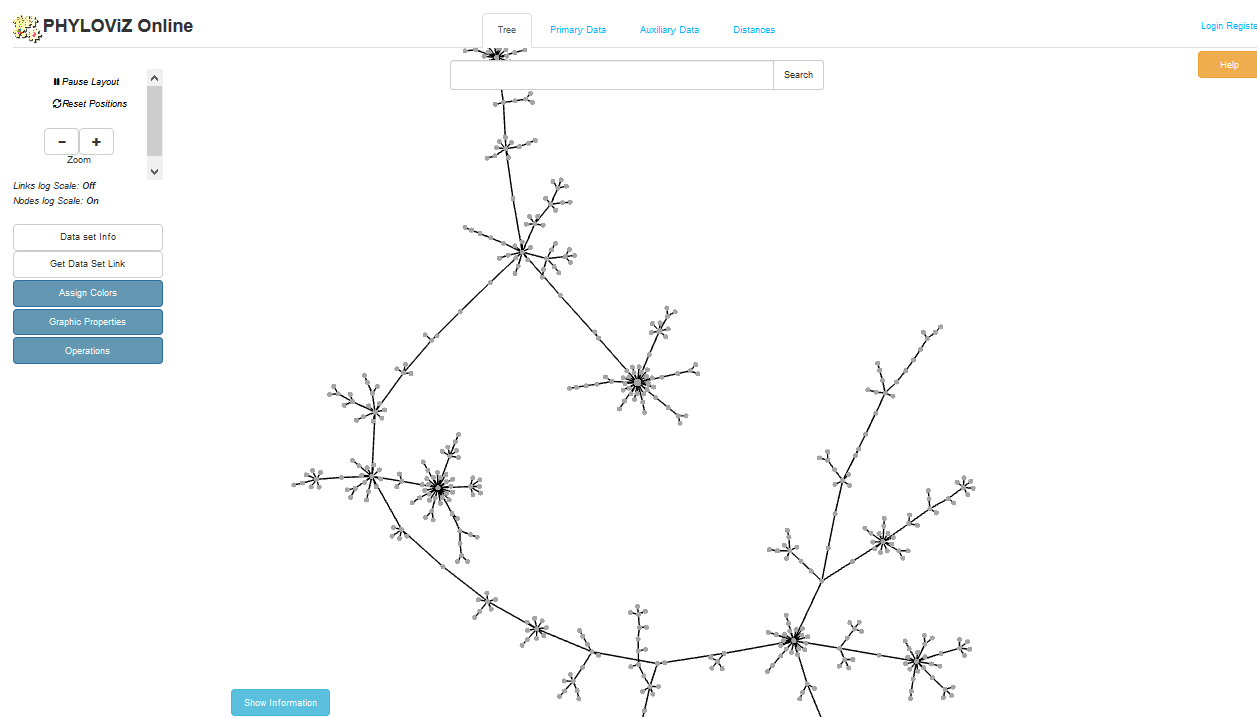
See more information about manipulating the tree on the PhyloViz website.