User-configurable options
The BIGSdb user interface is configurable in a number of ways. Choices made are remembered between sessions. If the database requires you to log on, the options are associated with your user account, whereas if it is a public database, that you haven’t logged in to, the options are associated with a browser cookie so they will be remembered if you connect from the same computer (using the same browser).
Most options are set by clicking the ‘Customise’ link on the database contents page. Most of the available options are visible for isolate databases, whereas sequence definition databases have fewer available.
General options
The general options tab is displayed by default. If another tab is being shown, click the ‘General options’ header.
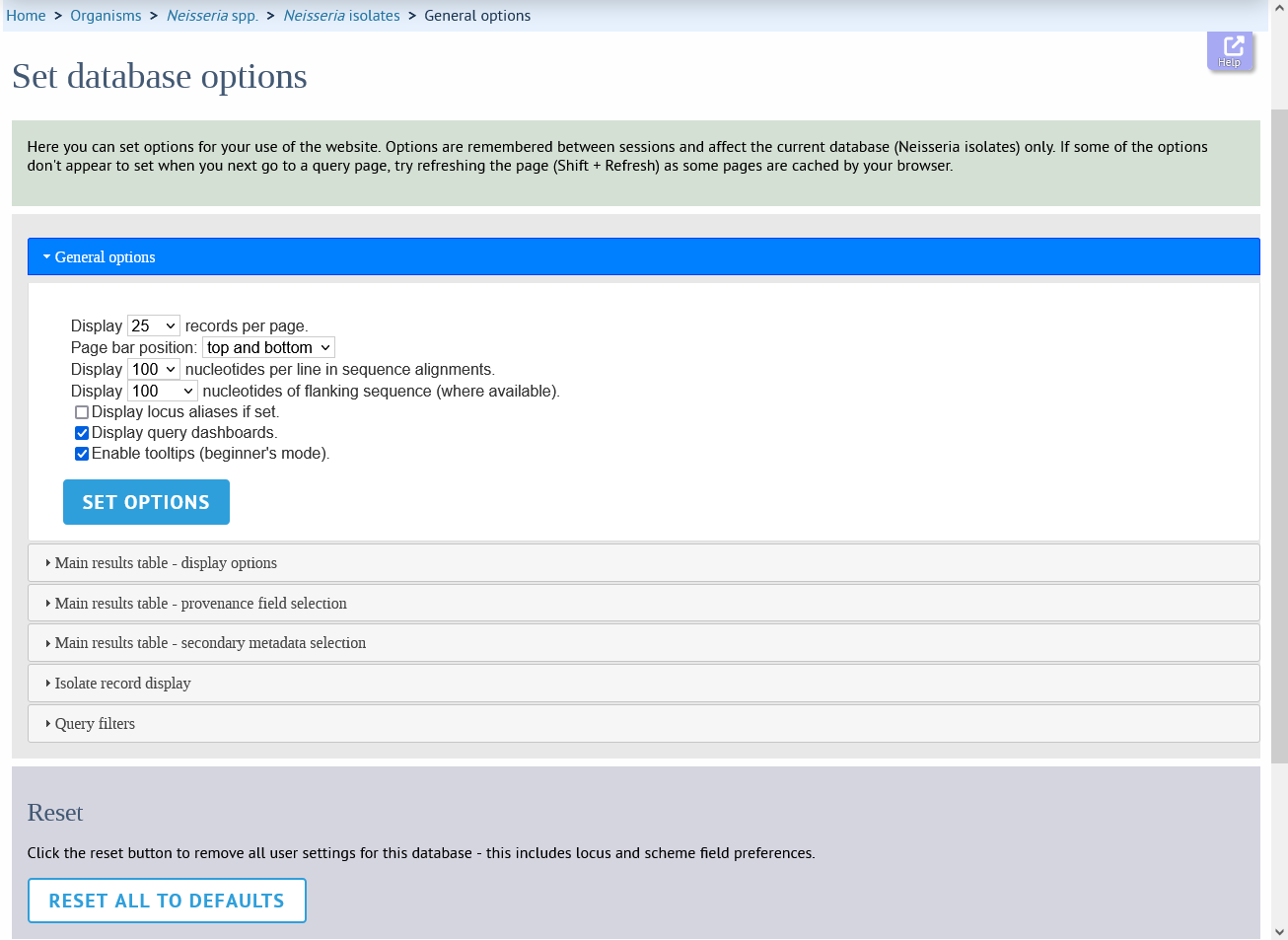
The general tab allows the following options to be modified:
Records per page
Page bar position
Nucleotides per line - Some analyses display sequence alignments. This option allows you to set the width of these alignments so suit your display.
Flanking sequence length - This sets the length of flanking sequence upstream and downstream of a particular locus that is included whenever a sequence is displayed. Flanking sequences are displayed fainter that the locus sequence.
Locus aliases - Loci can have multiple names (aliases). Setting this option will display all alternative names in results tables.
Tooltips (beginner’s mode) - Most query forms have help available in the form of information tooltips. These can be switched on/off here. They can also be toggled off by clicking the Toggle: ‘i’ button at the top-right of the display of some pages.
Click ‘Set options’ to remember any changes you make.
Main results table
The ‘main results table’ tab contains options for the display of paged results following a query.
Click the ‘Main results table’ header to display the tab.
The ‘main results table’ tab will scroll up.
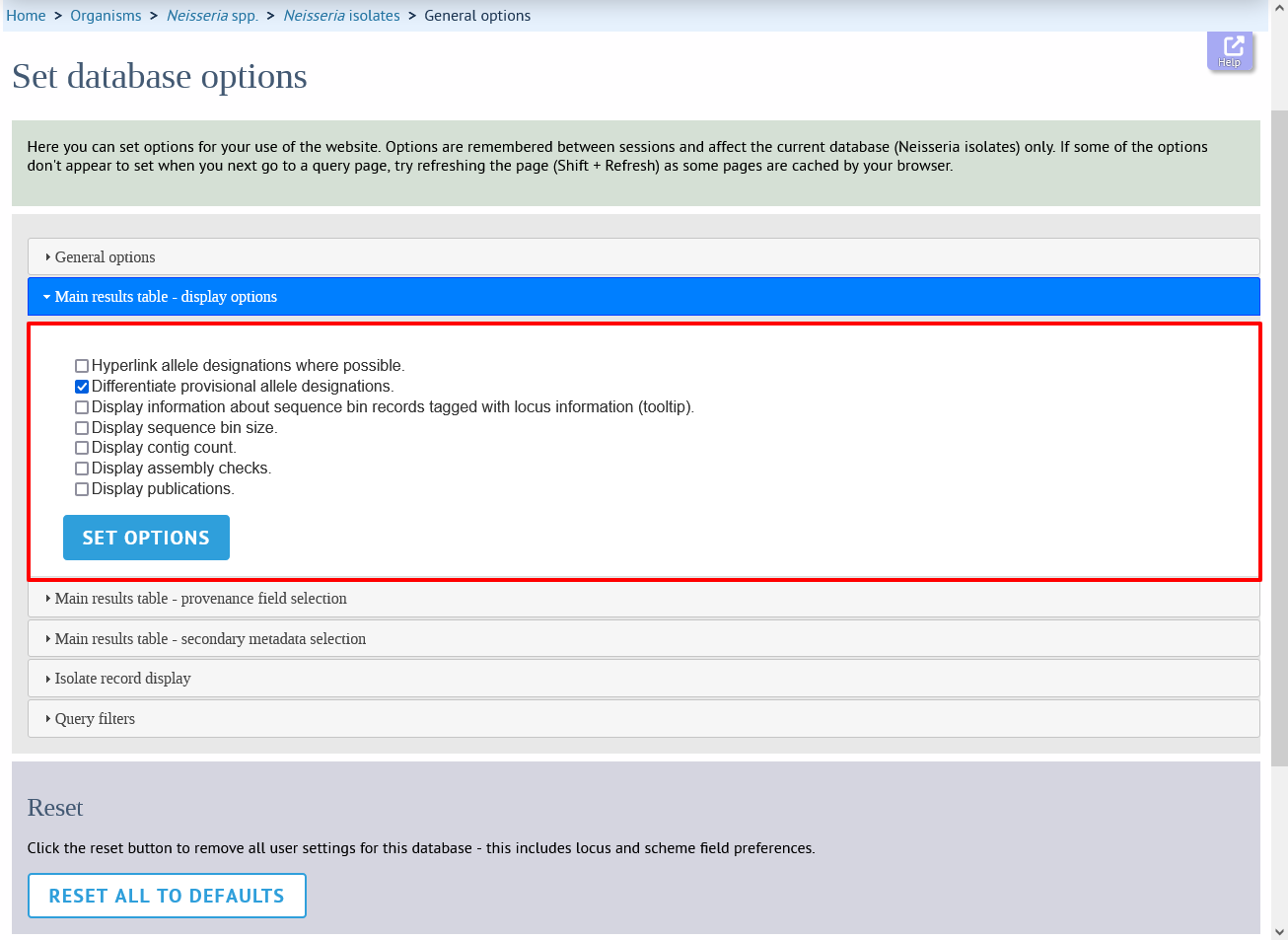
This tab allows the following options to be modified:
Hyperlink allele designations - Hyperlinks point to an information page about the particular allele definition. Depending on the locus, these may exist on a different website.
Differentiate provisional allele designations - Allele designations can be set as confirmed or provisional, usually depending on the method of assignment. Selecting this option will display provisional designations in a different colour to confirmed designations.
Information about sequence bin records - Creates a tooltip that displays details about sequence tags corresponding to a locus.
Sequence bin records - Displays a tooltip linking to the sequence tag if available.
Sequence bin size - Displays the size of the sum of all contigs associated with each isolate record.
Contig count - Displays the number of contigs associated with each isolate record.
Assembly checks - Displays whether assembly checks have passed for a genome record.
Publications - Displays citations with links to PubMed for each record.
Isolate record display
The ‘isolate record display’ tab contains options for the display of a full isolate record.
Click the ‘Isolate record display’ tab to display the tab.
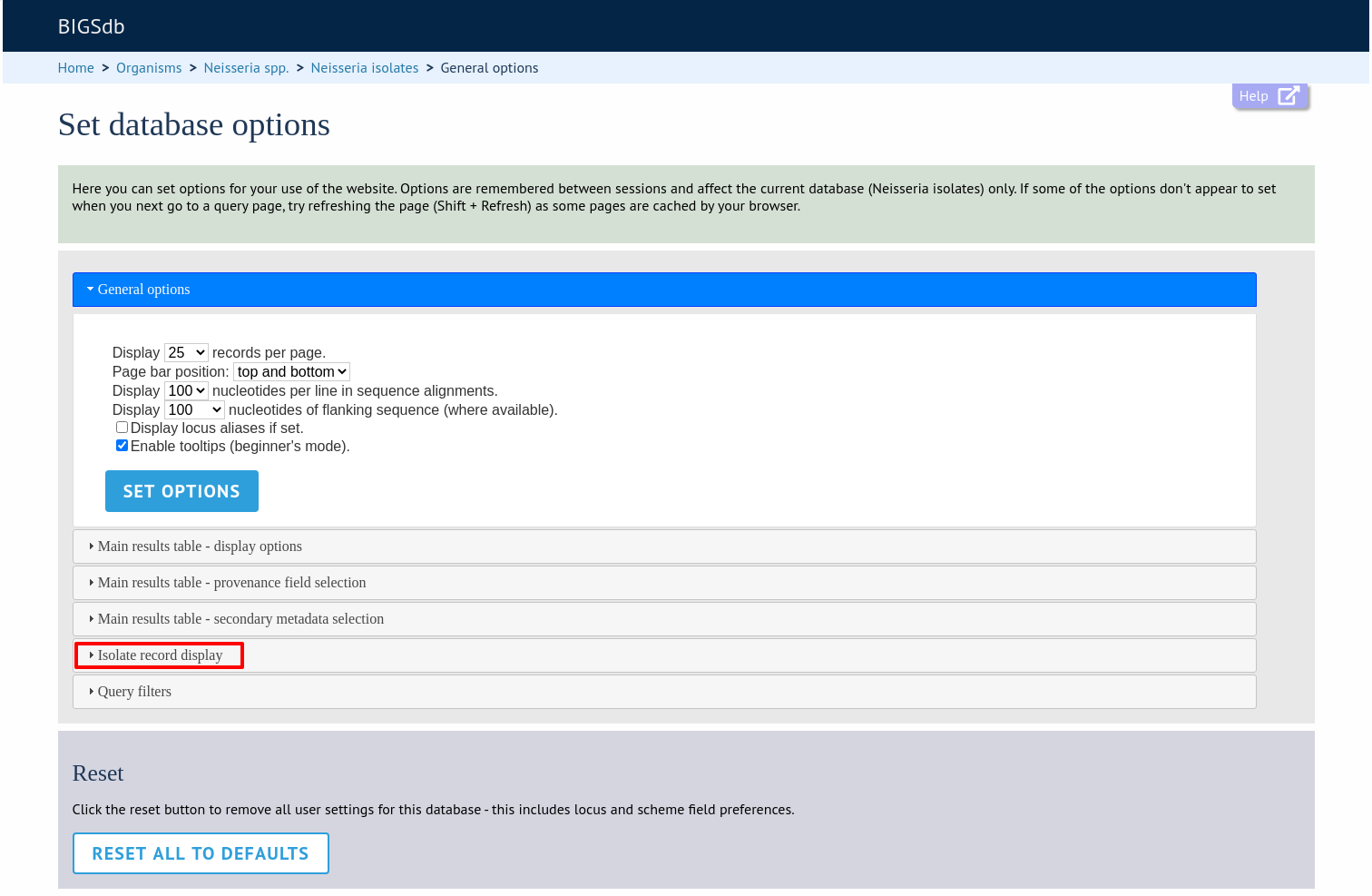
The ‘Isolate record display’ tab will scroll up.
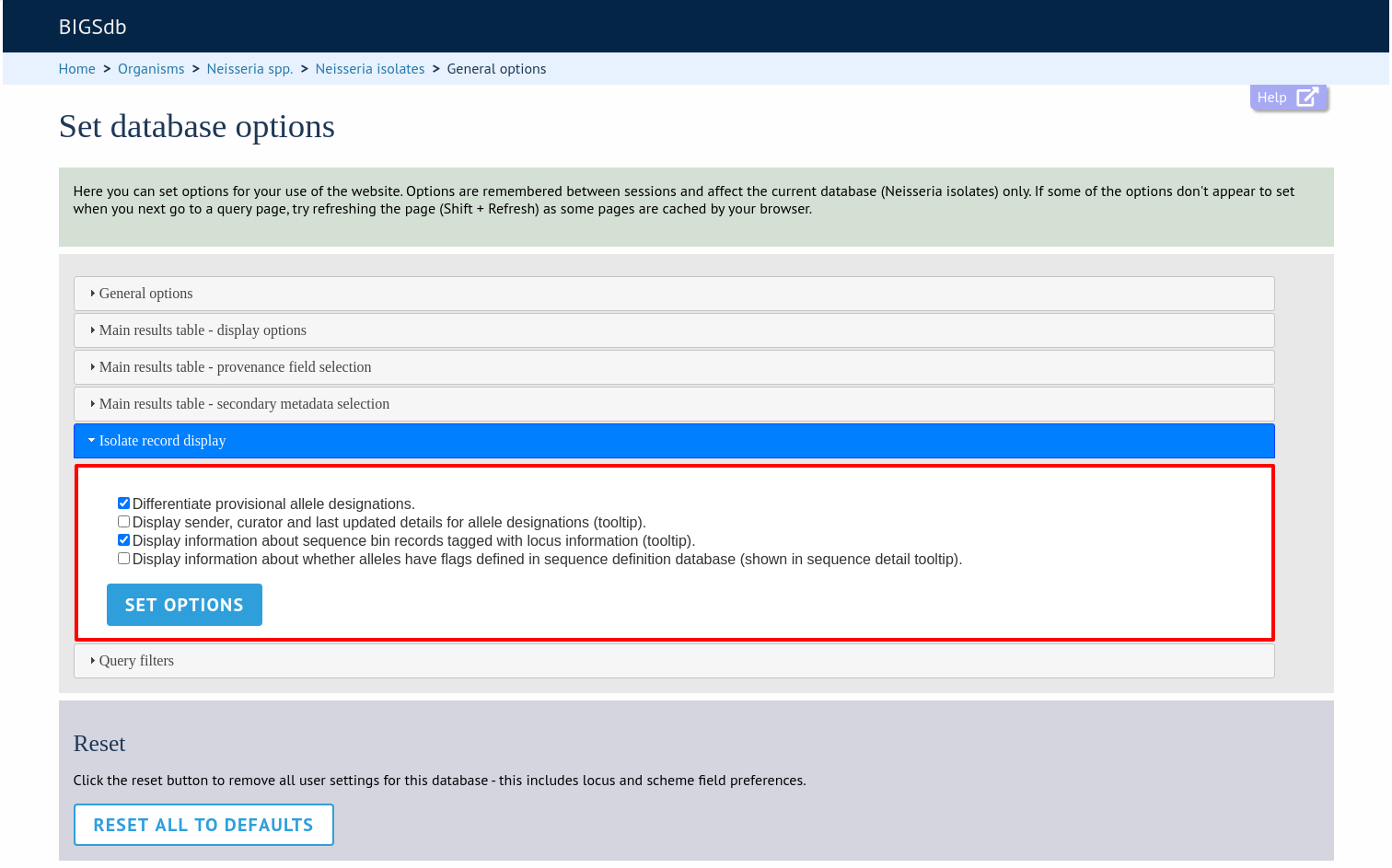
This tab allows the following options to be modified:
Differentiate provisional allele designations - Allele designations can be set as confirmed or provisional, usually depending on the method of assignment. Selecting this option will display provisional designations in a different colour to confirmed designations.
Display sender, curator and last updated records - Displays a tooltip containing sender information next to each allele designation.
Sequence bin information - Displays a tooltip with information about the position of the sequence if tagged within the sequence bin.
Allele flags - Displays information about whether alleles have flags defined in sequence definition databases.
Provenance field display
The ‘provenance field display’ tab contains checkboxes for fields to display in the main results table.
Click the ‘Provenance field display’ tab to display the tab.
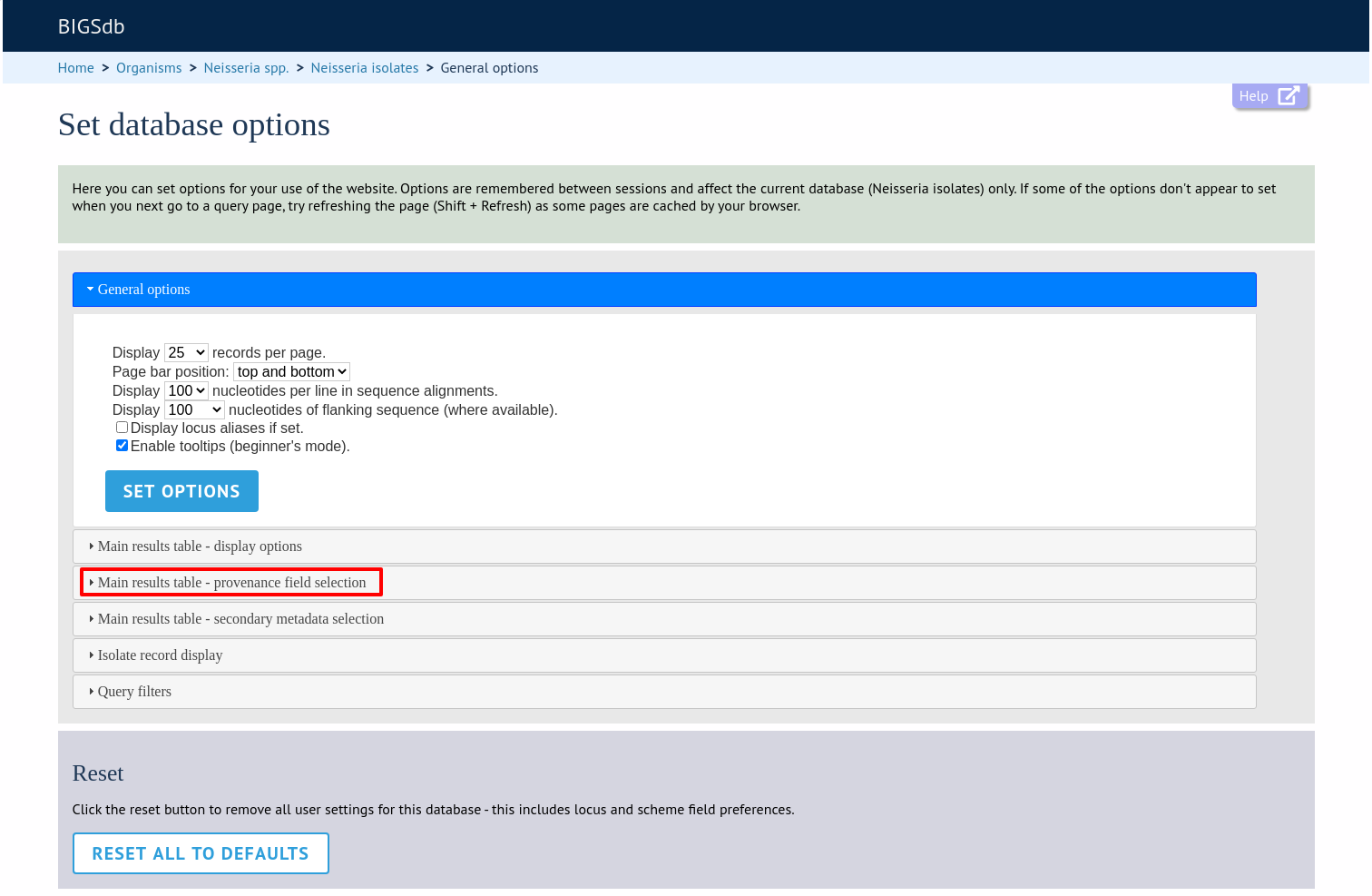
The ‘Provenance field display’ tab will scroll up.
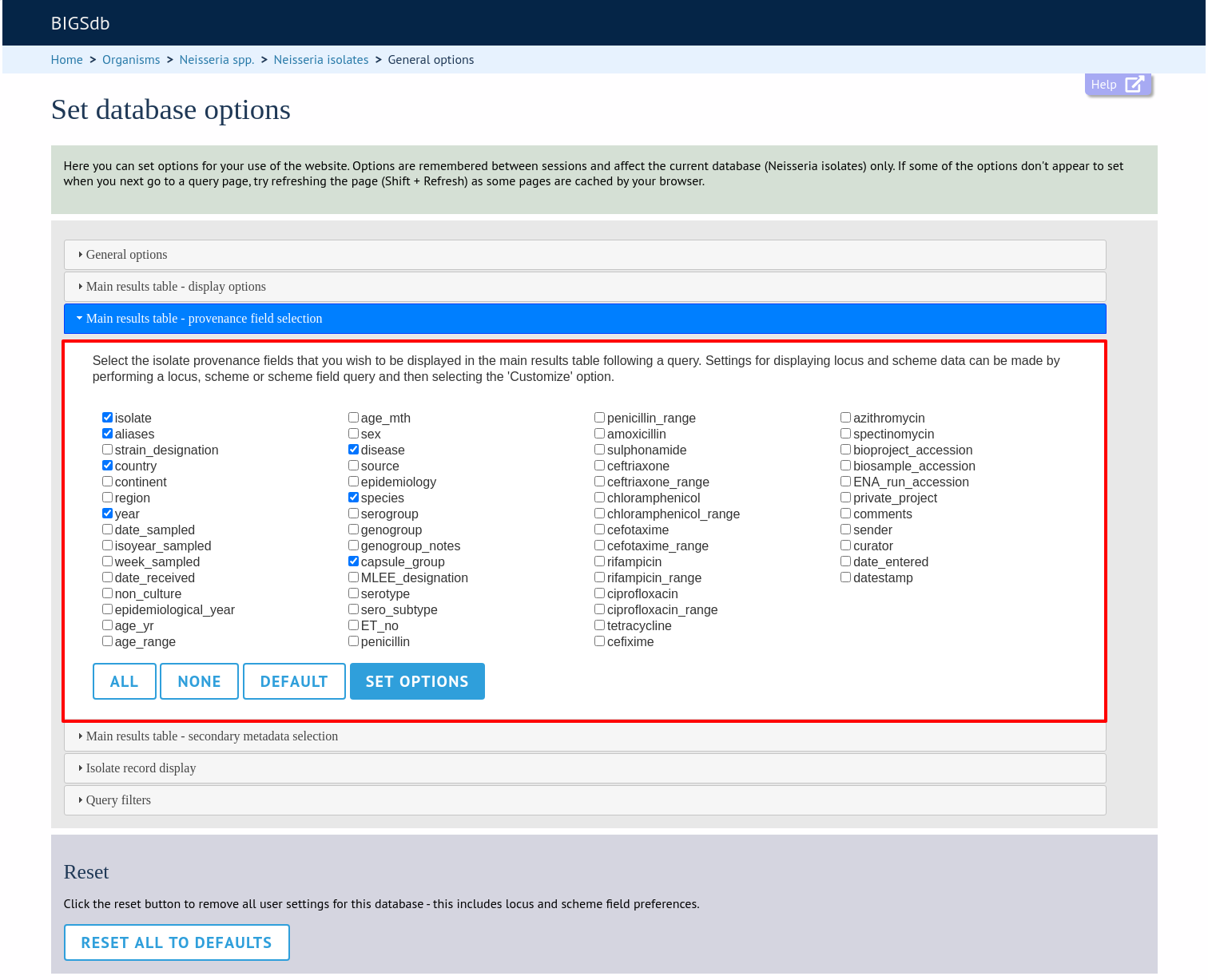
Some fields will be checked by default - these are defined during database setup (maindisplay option).
Check any fields that you wish to be displayed and then click ‘Set options’. You can return to the default selection by clicking ‘Default’ followed by ‘Set options’.
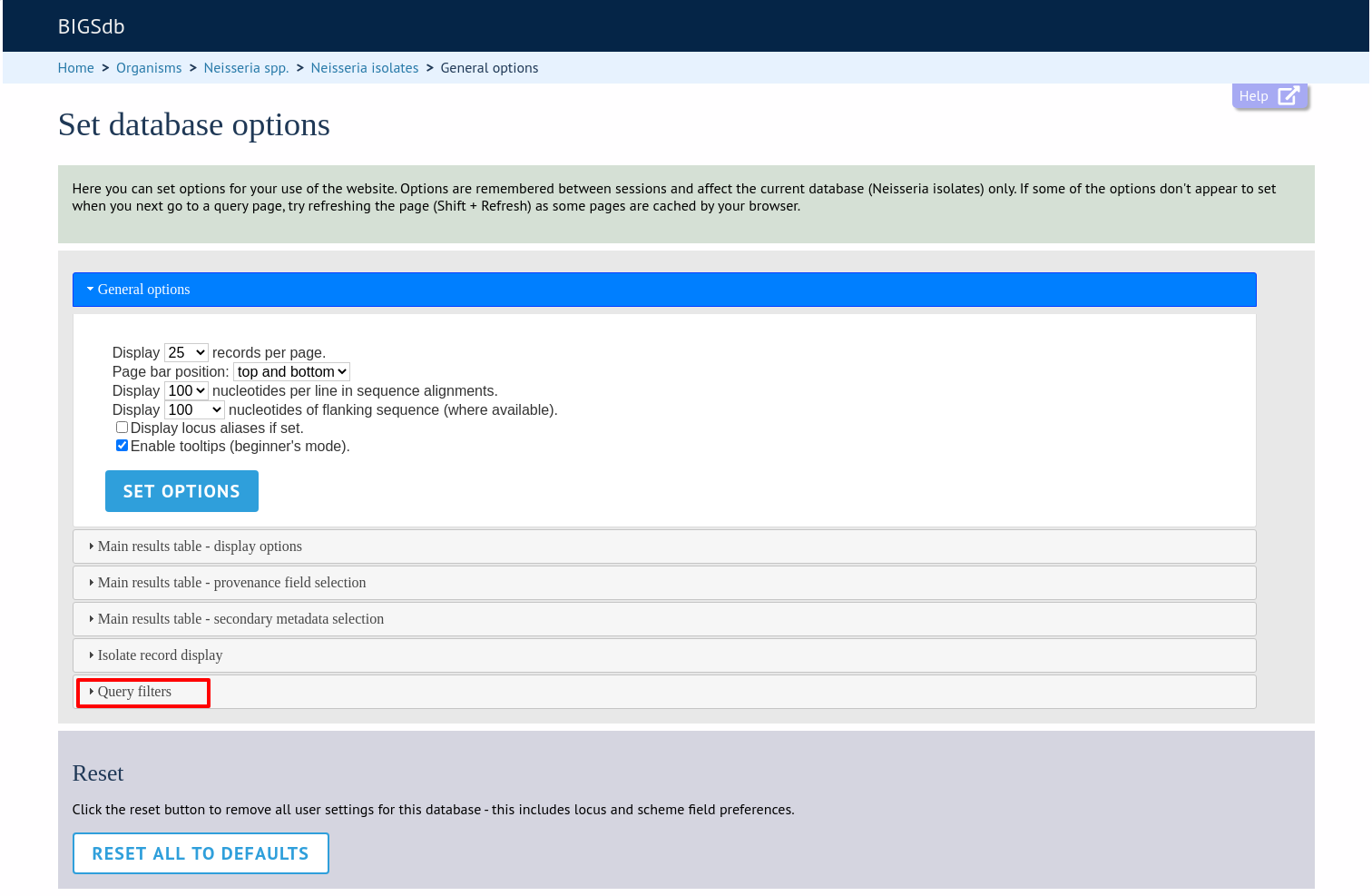
Query filters
The ‘query filters’ tab contains checkboxes for provenance fields and scheme completion status. Checking these results in drop-down list box filters appearing in the query page filters fieldset.
Click the ‘Query filters’ tab to display the tab.
The ‘Query filters’ tab will scroll up.
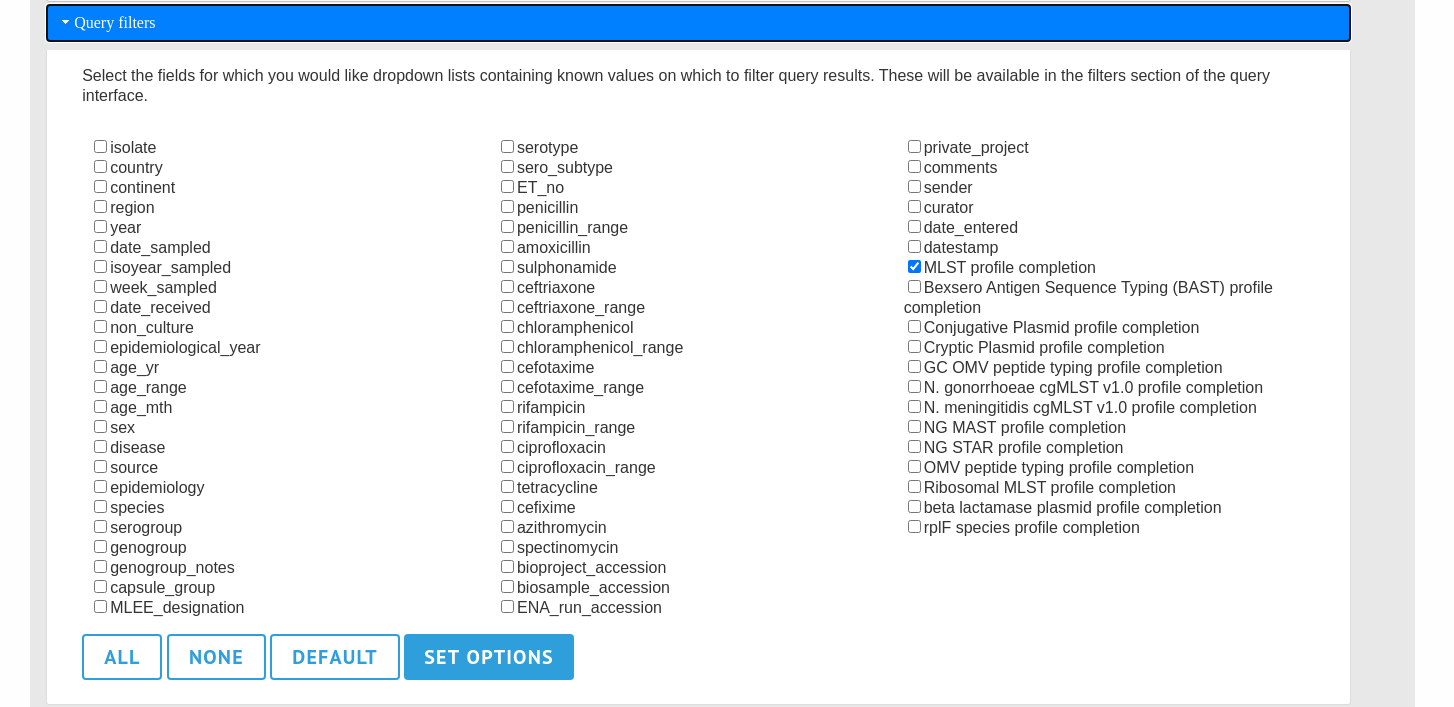
A list of possible filters appears. Click any checkbox for a filter you would like to make available. Click ‘Set options’ when done. You can return to the default selection by clicking ‘Default’ followed by ‘Set options’.
Modifying locus and scheme display options
Whether or not loci, schemes or scheme fields are displayed in result tables, isolate records, or within query dropdown boxes can all be set with default options when first defined. These attributes can, however, be overridden by a user, and these selections will be remembered between sessions.
The procedure to modify these attributes is the same for locus, schemes or scheme fields, so the steps for loci will be demonstrated only.
Click the appropriate link in the ‘Customise’ section on the isolate contents page.
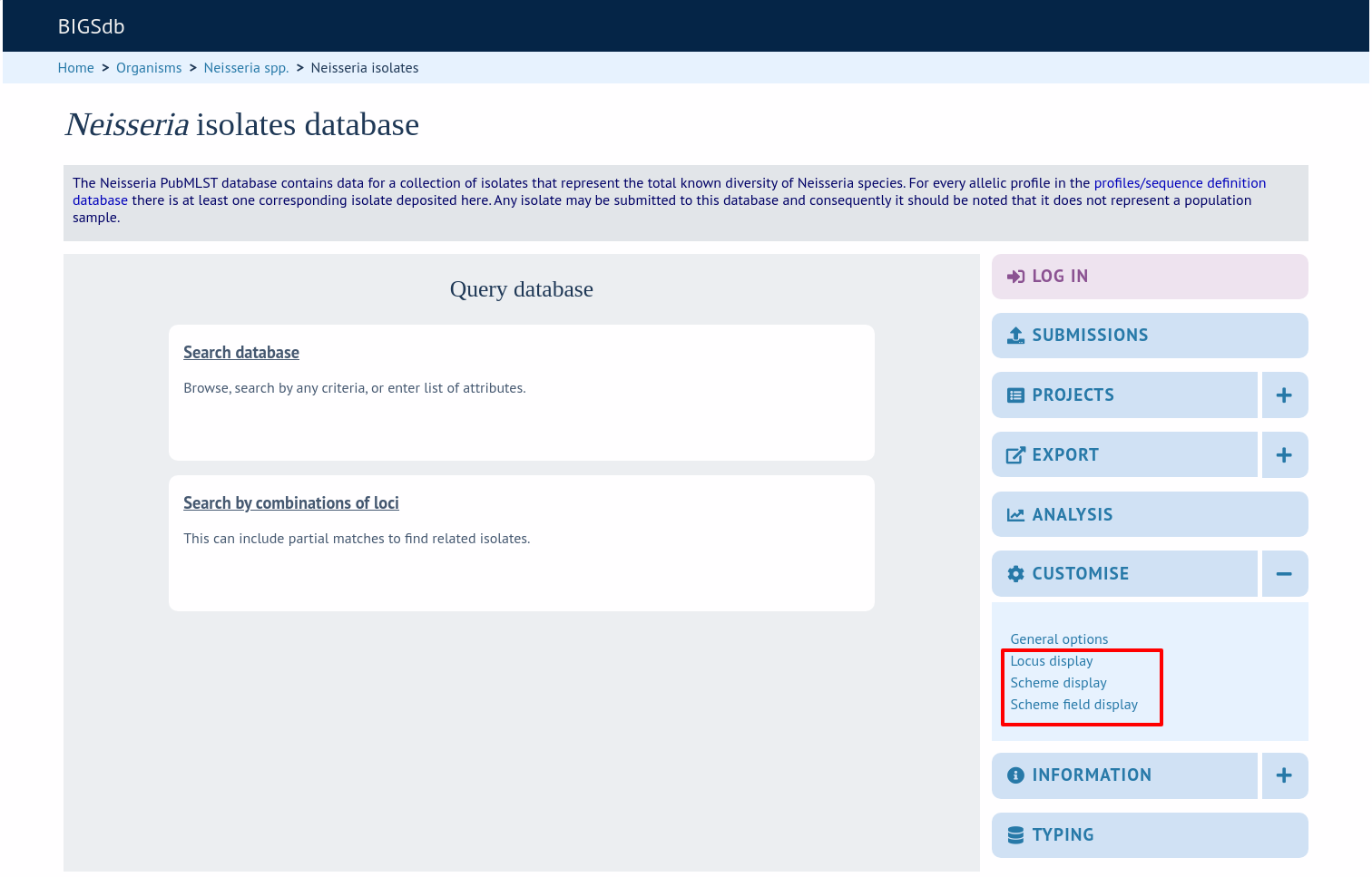
Either select the locus id by querying for it directly.
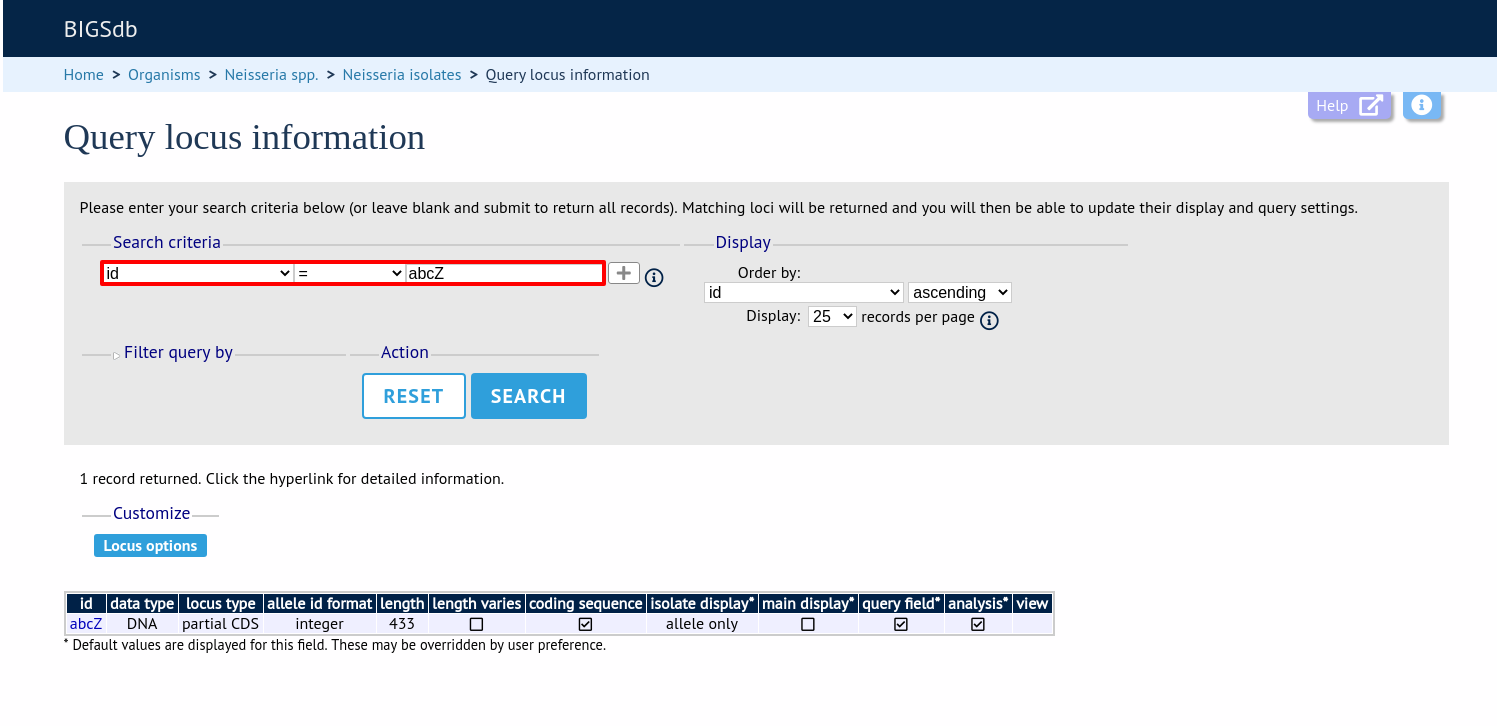
Designations can be queried using standard operators.
Alternatively, you can search by filtering loci by schemes. Click the ‘Filter query by’ header and select the scheme in the dropdown box.
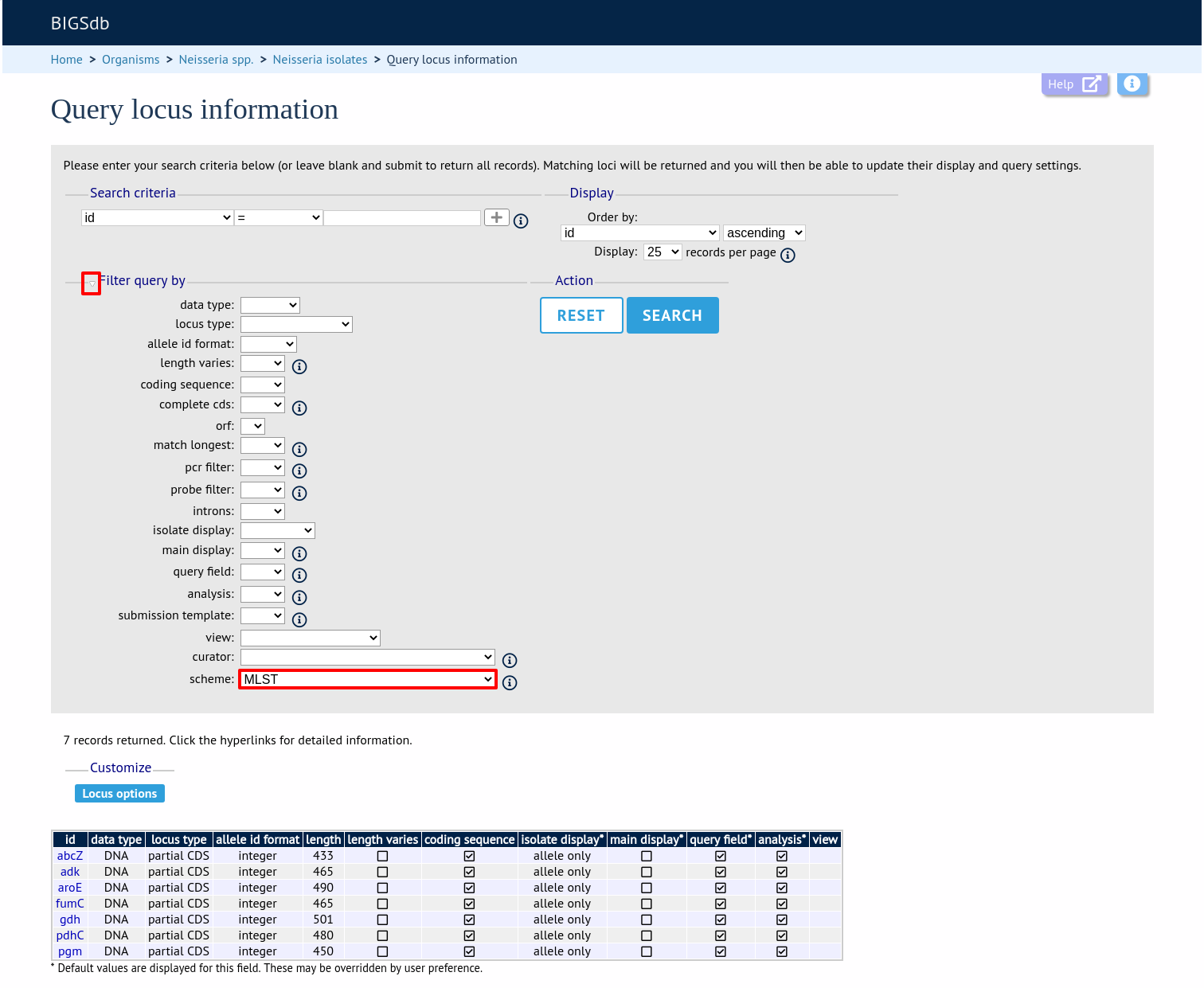
Once loci have been selected, click Customize ‘locus options’.
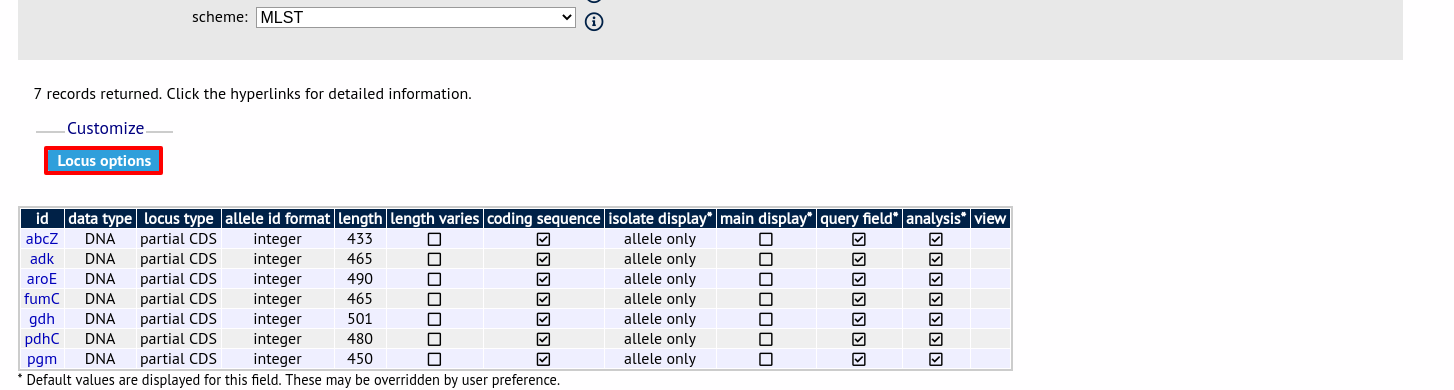
You can then choose to add or remove individual loci from the selection by clicking the appropriate checkboxes. At the bottom of the page are a number of attributes that you can change - clicking ‘Change’ will affect all selected loci.
Possible options for loci are:
isolate_display - Sets how the locus is displayed within an isolate record:
allele only - display only identifier
sequence - display the full sequence
hide - don’t show at all
main_display - Sets whether the locus is displayed in the main results table following a query.
query_field - Sets whether the locus appears in dropdown list boxes to be used within queries.
analysis - Sets whether the locus can be used in data analysis functions.
Note
Settings for loci can be overridden by those set for schemes that they are members of. For example, if you set a locus to be displayed within a main results table, but that locus is a member of a scheme and you set that scheme not to be displayed, then the locus will not be shown. Conversely, if you set a scheme to be displayed, but set its member locus not to be shown, then that locus will not be displayed (but other loci and scheme fields may be, depending on their independent settings).