Locus explorer
The locus explorer is a sequence definition database plugin. It can create schematics showing the polymorphic sites within a locus, calculate the GC content and generate aligned translated sequences.
Expand the ‘Analysis’ section on the main contents page of a sequence definition database and click ‘Locus Explorer’.
Polymorphic site analysis
Select the locus you would like to analyse in the Locus dropdown box. The page will reload.
Select the alleles that you would like to include in the analysis. Variable length loci are limited to 2000 sequences or fewer since these need to be aligned. Select ‘Polymorphic Sites’ in the Analysis selection and click ‘Submit’.
If an alignment is necessary, the job will be submitted to the job queue and the analysis performed. If no alignment is necessary, then the analysis is shown immediately.
The first part of the page shows the schematic.
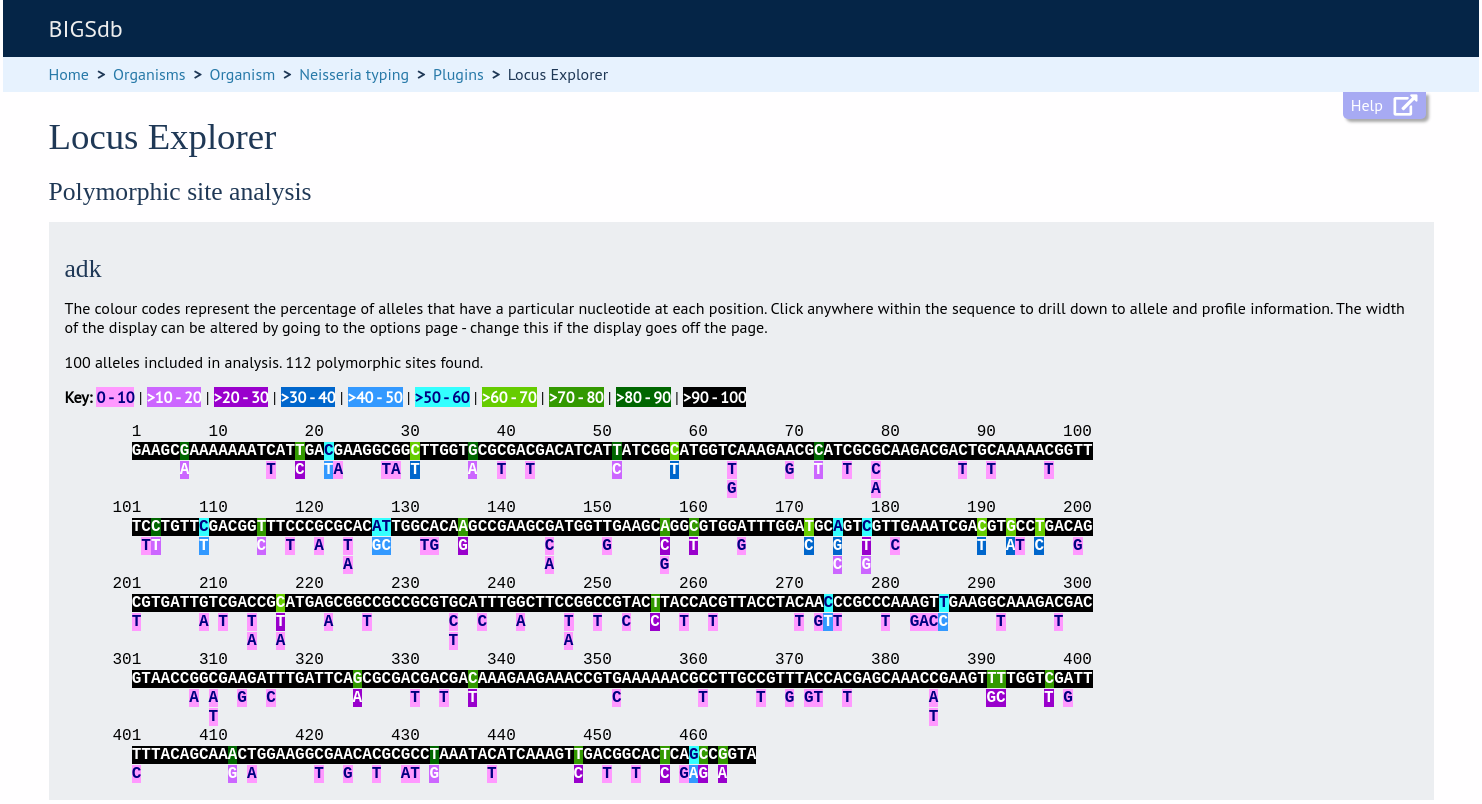
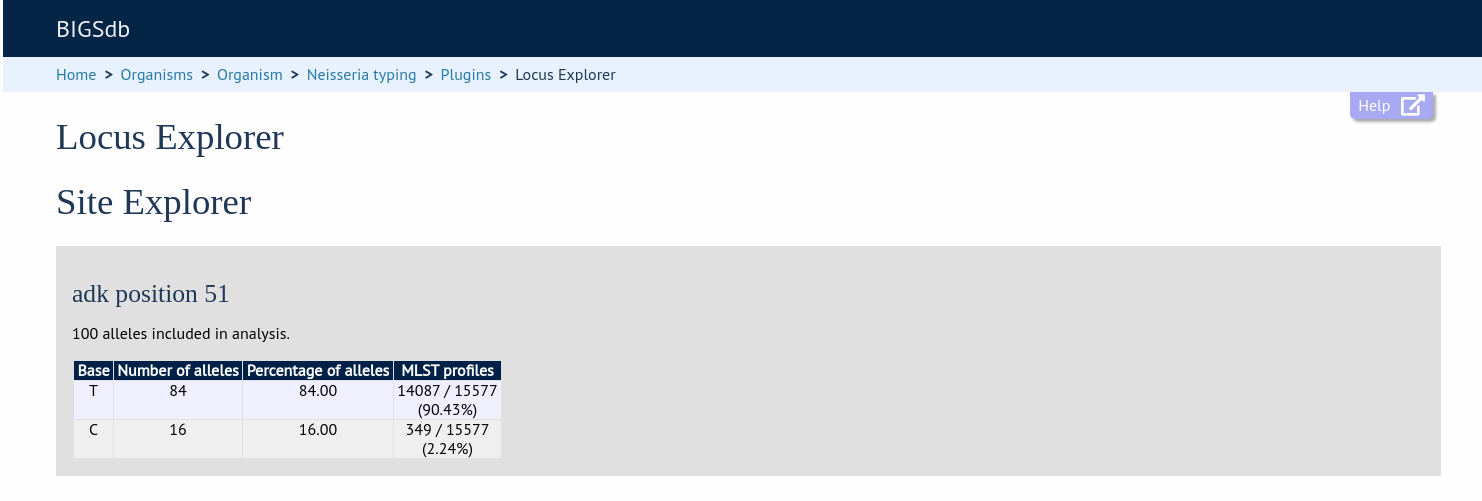
Clicking any of the sequence bases will calculate the exact frequencies of the different nucleotides at that position.
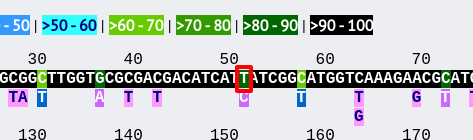
Along with the nucleotide frequencies, it will also show the percentage of allelic profiles containing each nucleotide at that position if the locus is part of a scheme such as MLST.
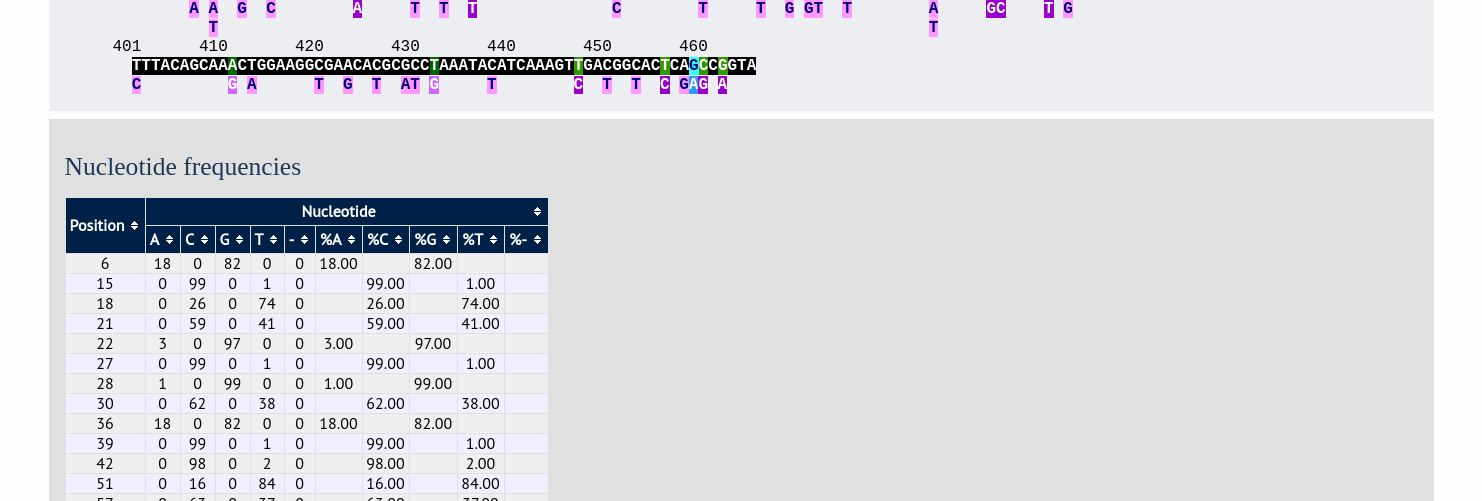
The second part of the page shows a table listing nucleotide frequencies at each of the variable positions.
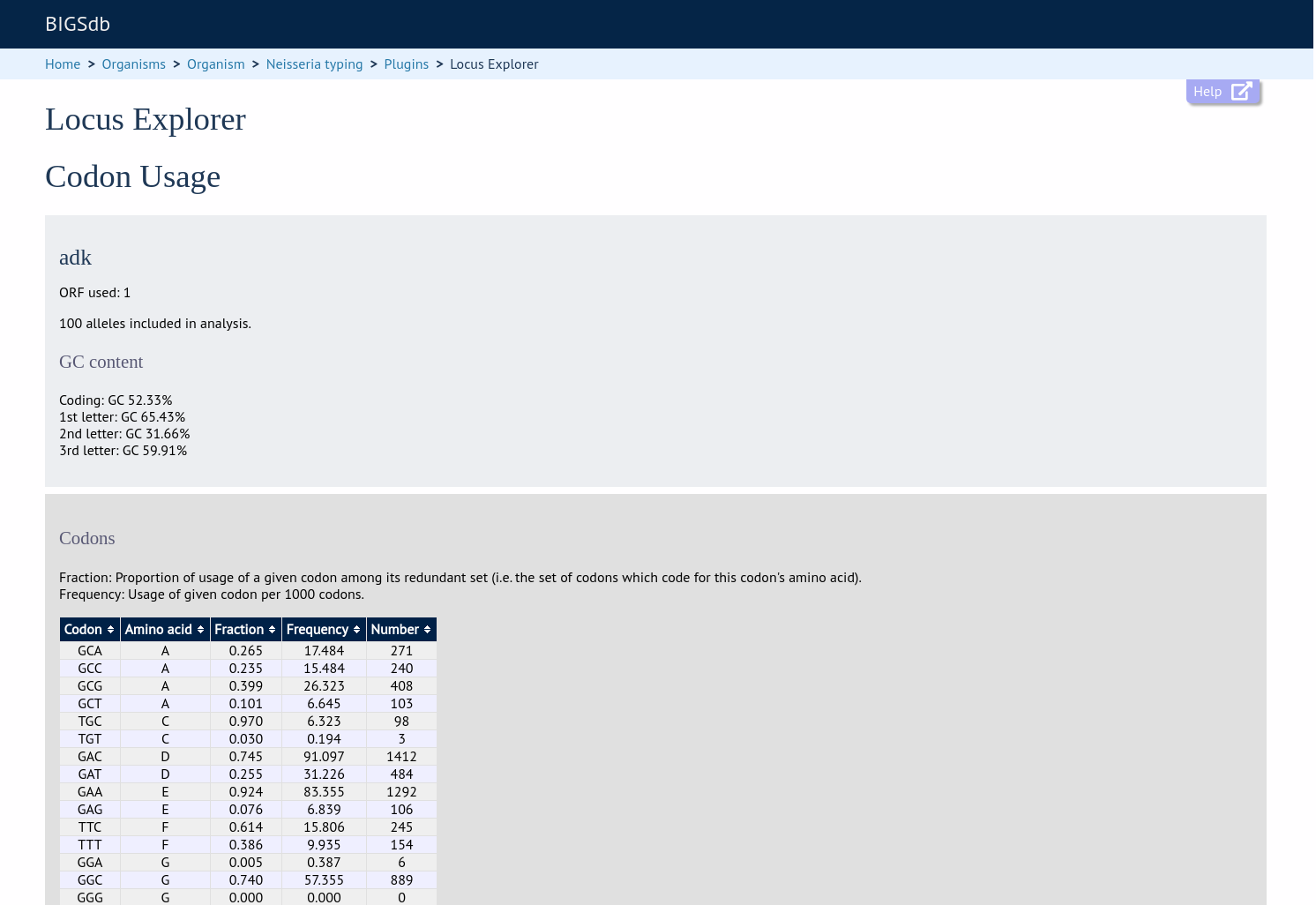
Codon usage
Select the alleles that you would like to include in the analysis. Again, variable length loci are limited to 200 sequences or fewer since these need to be aligned. Click ‘Codon’.
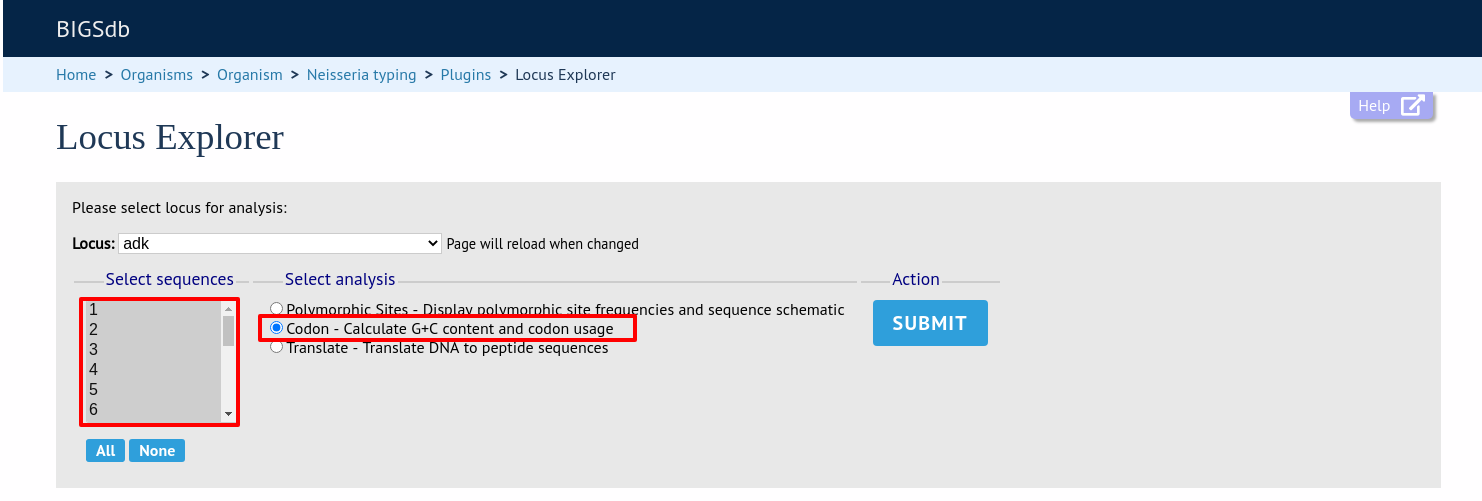
The GC content of the alleles will be determined and a table of the codon frequencies displayed.
Aligned translations
If a DNA coding sequence locus is selected, an aligned translation can be produced.
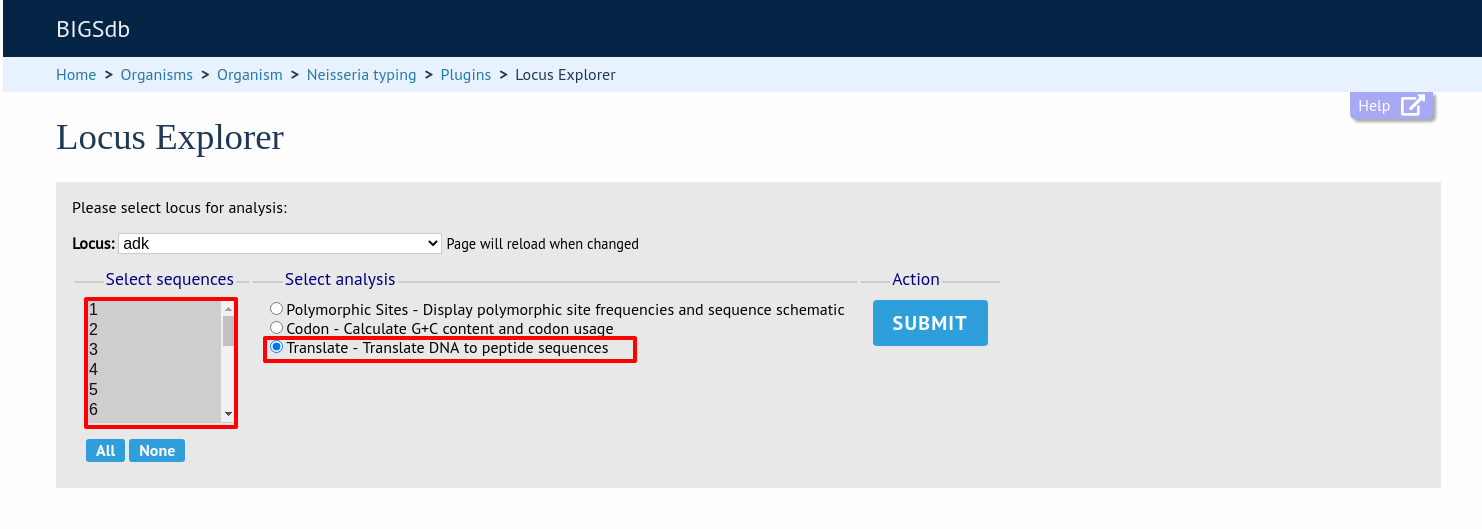
Select the alleles that you would like to include in the analysis. Again, variable length loci are limited to 200 sequences or fewer since these need to be aligned. Click ‘Translate’.
An aligned amino acid sequence will be displayed.
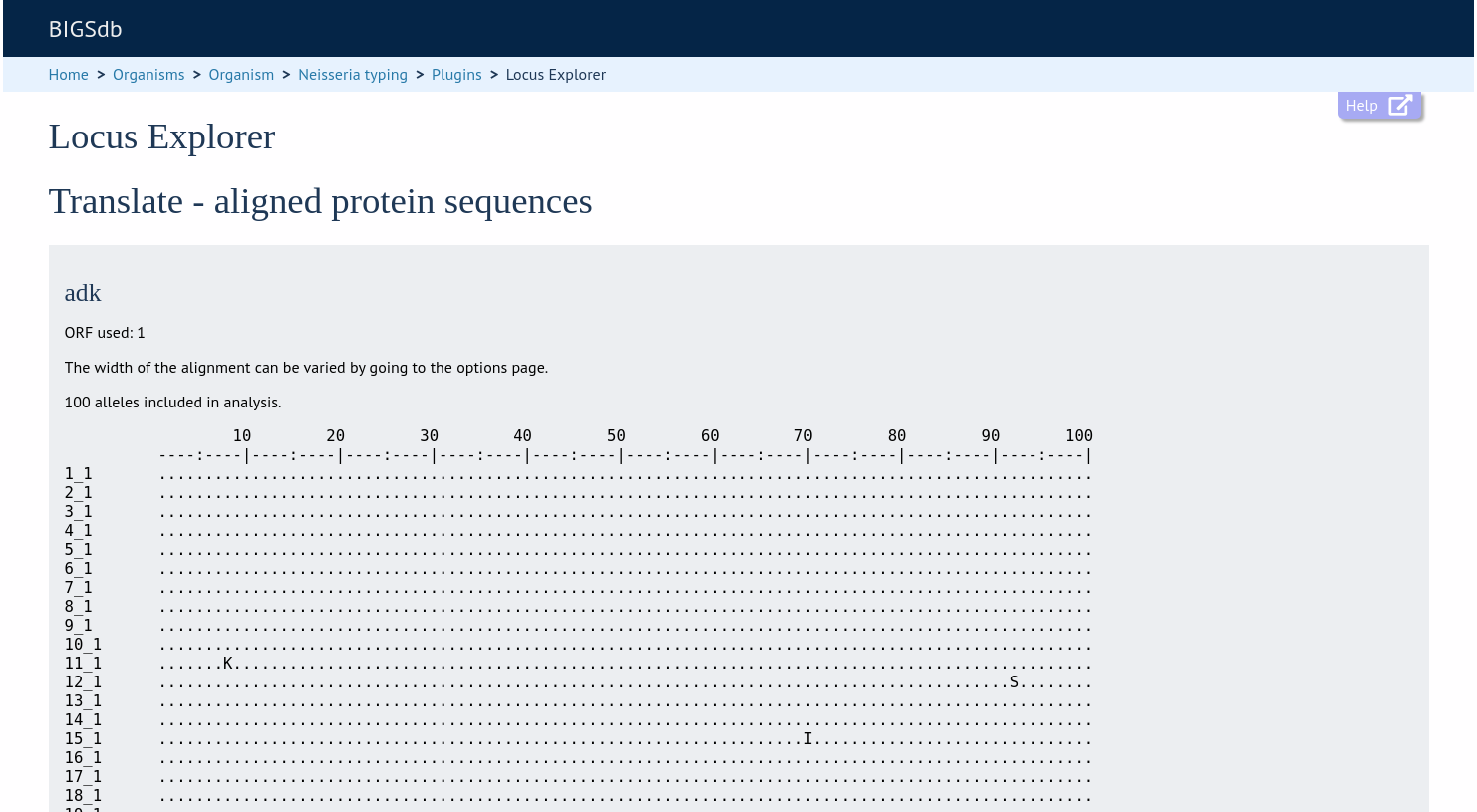
If there appear to be a lot of stop codons in the translation, it is possible that the orf value in the locus definition is not set correctly.