Retiring allele identifiers
Sometimes there is a requirement to prevent the automated assignment of a particular allele identifier - an allele with that identifier may have been commonly used and has since been removed. Reassignment of the identifier to a new sequence may lead to confusion, so in this instance, it would be better to prevent this.
You can retire an allele identifier by clicking the ‘Add’ retired allele ids link on the sequence database curators’ page. This function is normally hidden, so you may need to click the ‘Show all’ toggle to display it.
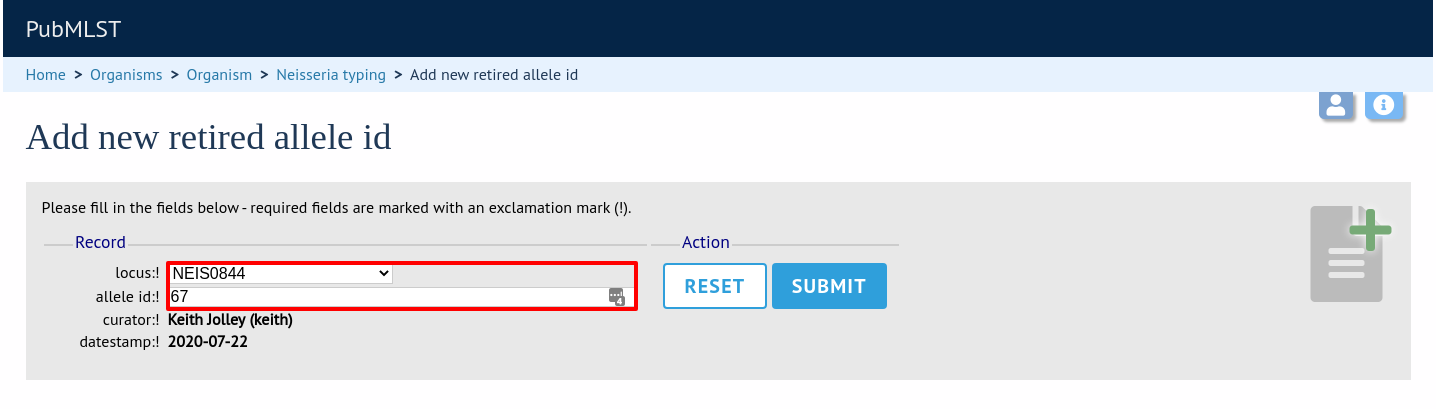
Select the locus from the dropdown list box and enter the allele id. Click ‘Submit’.
You cannot retire an allele that already exists, so you must delete it before retiring it. Once an identifier is retired, you will not be able to create a new allele with that name.
You can also retire an allele identifier when you delete an allele.
Un-retiring allele identifiers
If an allele identifier has been retired, it is possible to un-retire it so that it can be re-assigned. To do this, you need to remove the identifier from the retired_alleles table.
First, find the allele identifier in the retired_alleles table by clicking the ‘Update/delete’ retired alleles link on the sequence database curators’ page. This function is normally hidden, so you may need to click the ‘Show all’ toggle to display it.
Search by any criteria to find the allele identifier.
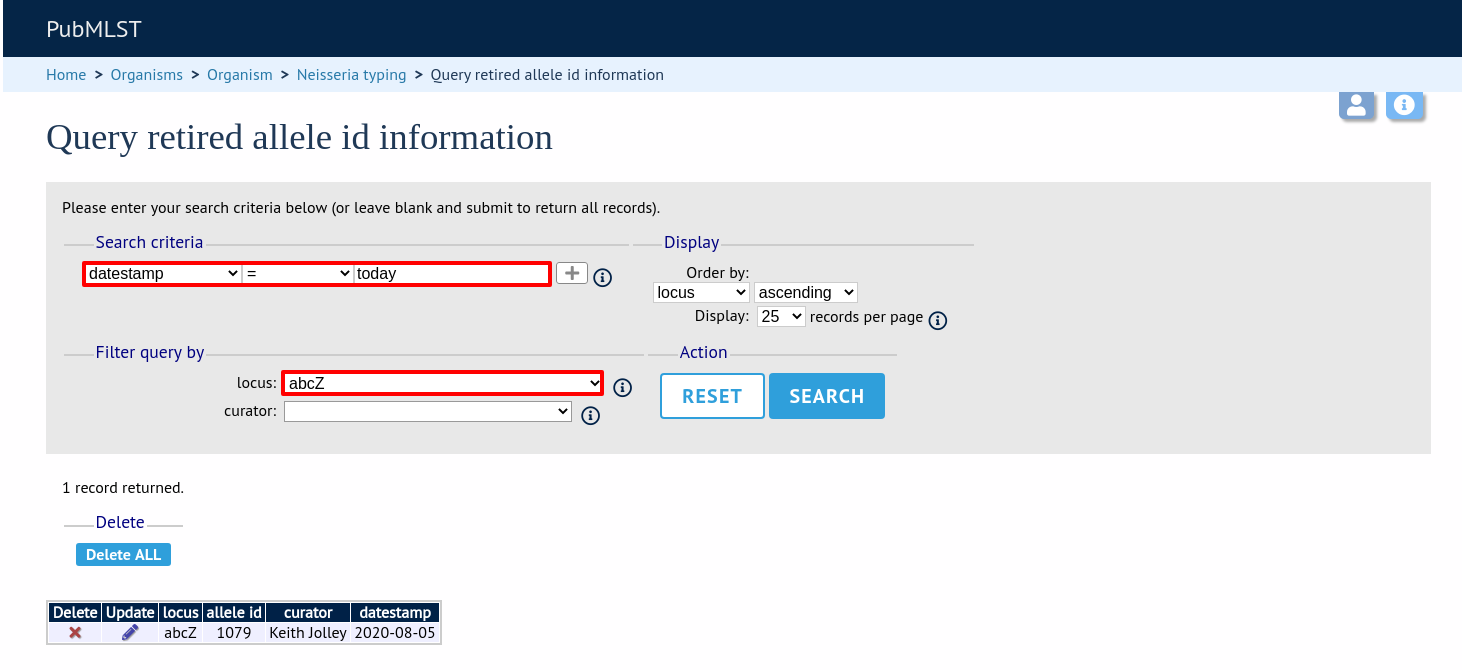
Click the delete link on the identifer to be un-retired.
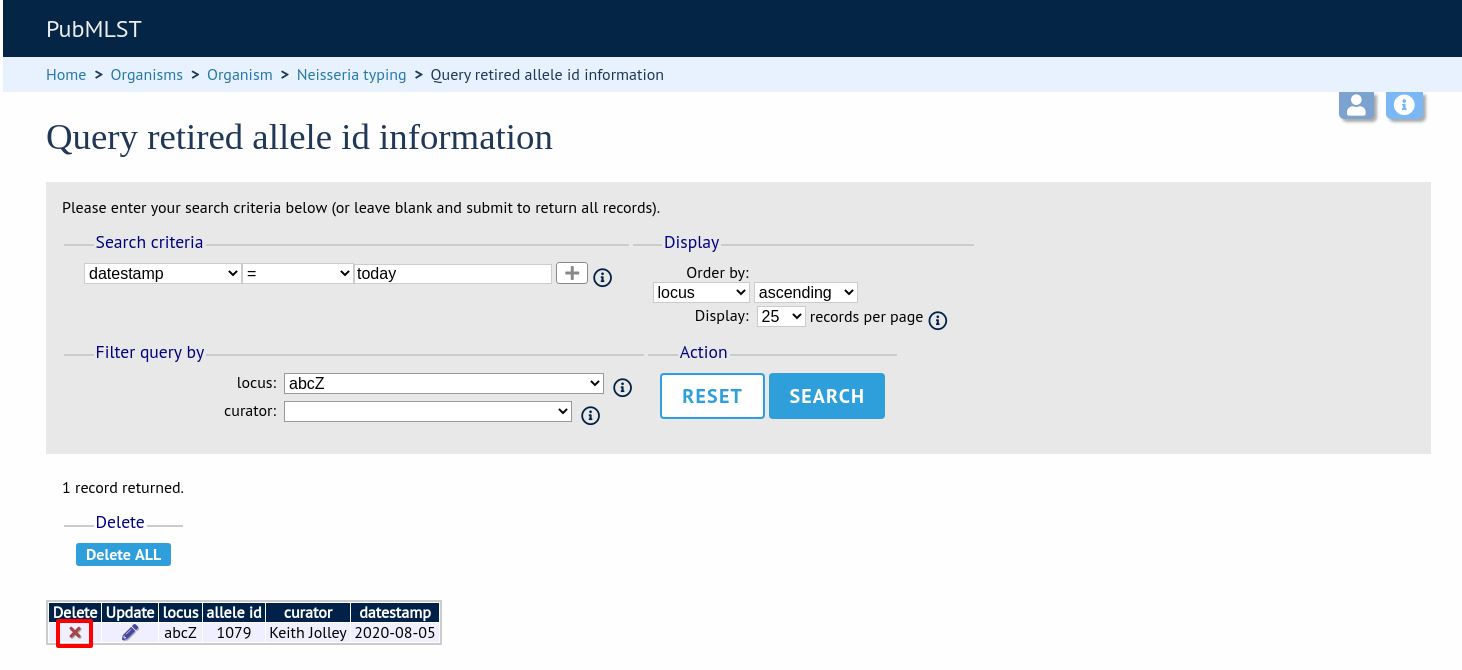
A confirmation page will be displayed. Click ‘Delete’ to remove the identifier from the retired alleles table.
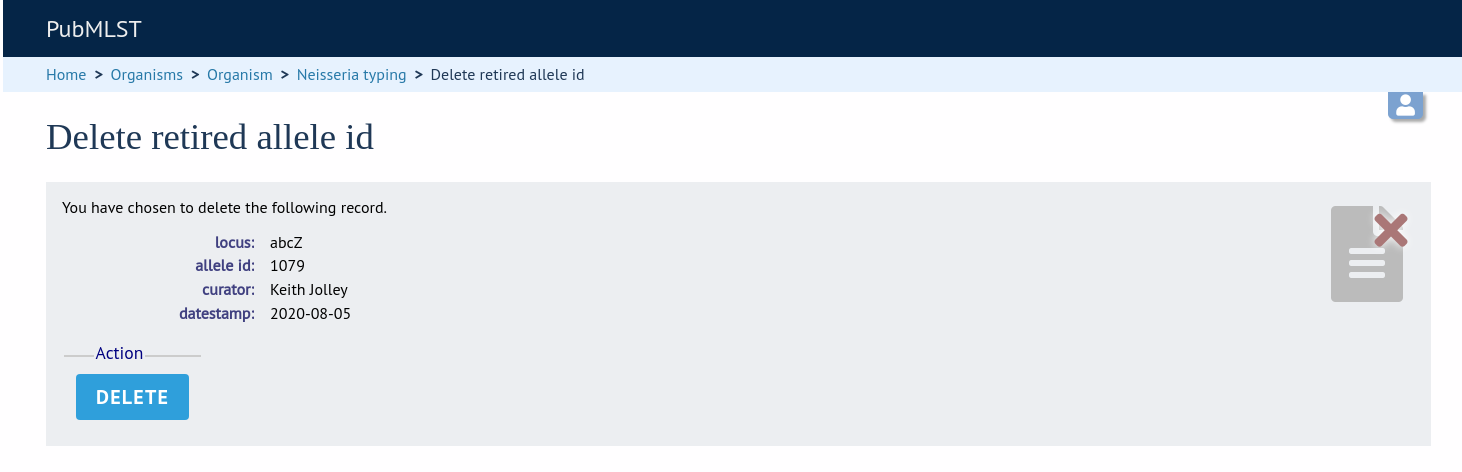
The identifier can now be re-assigned when adding a new allele.