Uploading sequence contigs linked to an isolate record
Select isolate from drop-down list
To upload sequence data, click the sequences add (+) sequence bin link on the curator’s main page.

Select the isolate that you wish to link the sequence to from the dropdown list box (or if the database is large and there are too many isolates to list, enter the id in the text box). You also need to enter the person who sent the data. Optionally, you can add the sequencing method used.
Paste sequence contigs in FASTA format in to the form.
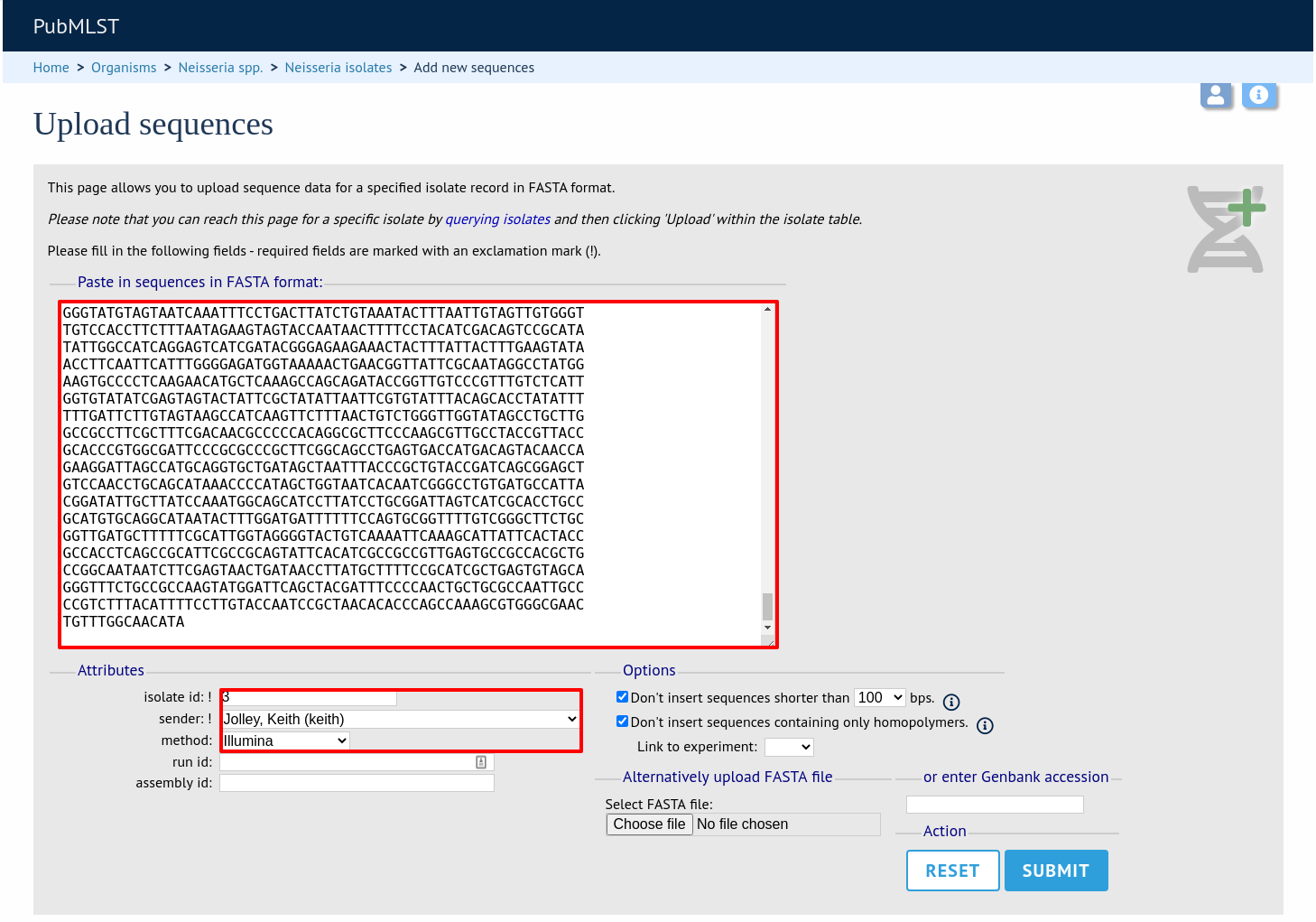
Click ‘Submit’. A summary of the number of isolates and their lengths will be displayed. To confirm upload, click ‘Upload’.
Select from isolate query
As an alternative to selecting the isolate from a dropdown list (or entering the id on large databases), it is also possible to upload sequence data following an isolate query.
Click the isolate update/delete link from the curator’s main page.

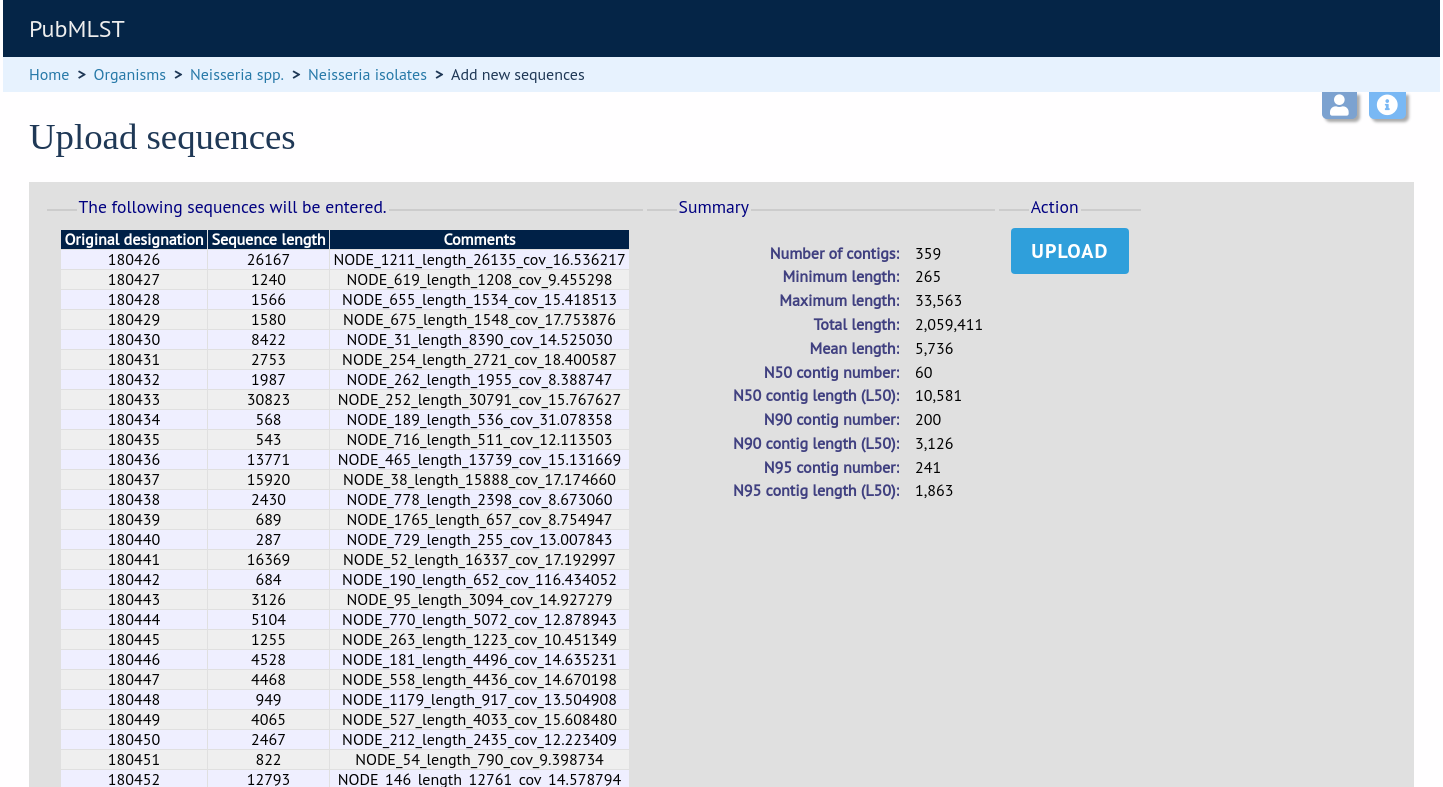
Enter your search criteria. From the list of isolates displayed, click the ‘Upload’ link in the sequence bin column of the appropriate isolate record.
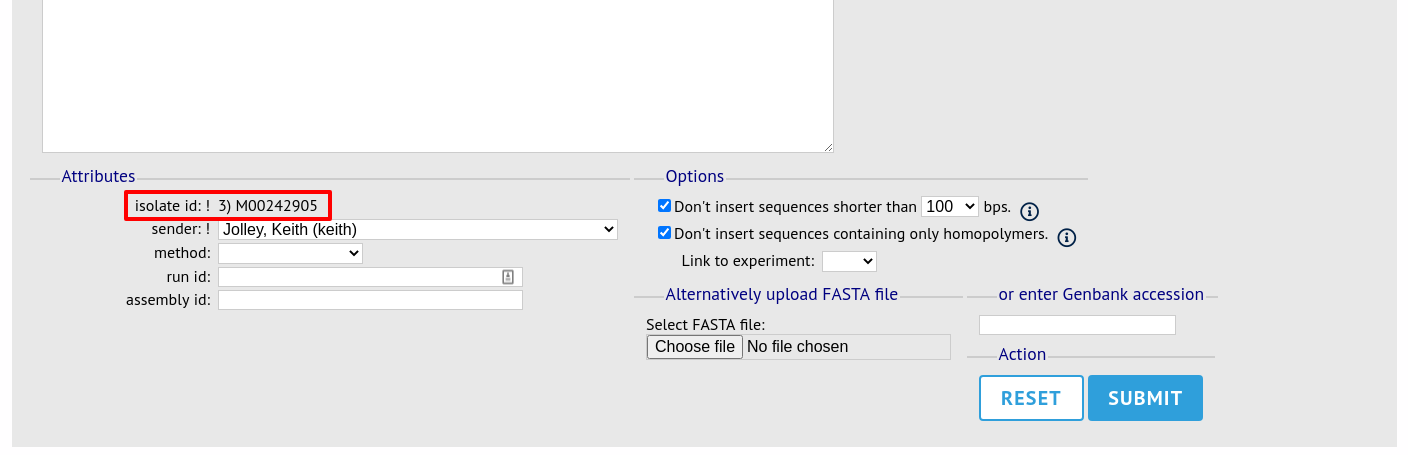
The same upload form as detailed above is shown. Instead of a dropdown list for isolate selection, however, the chosen isolate will be pre-selected.
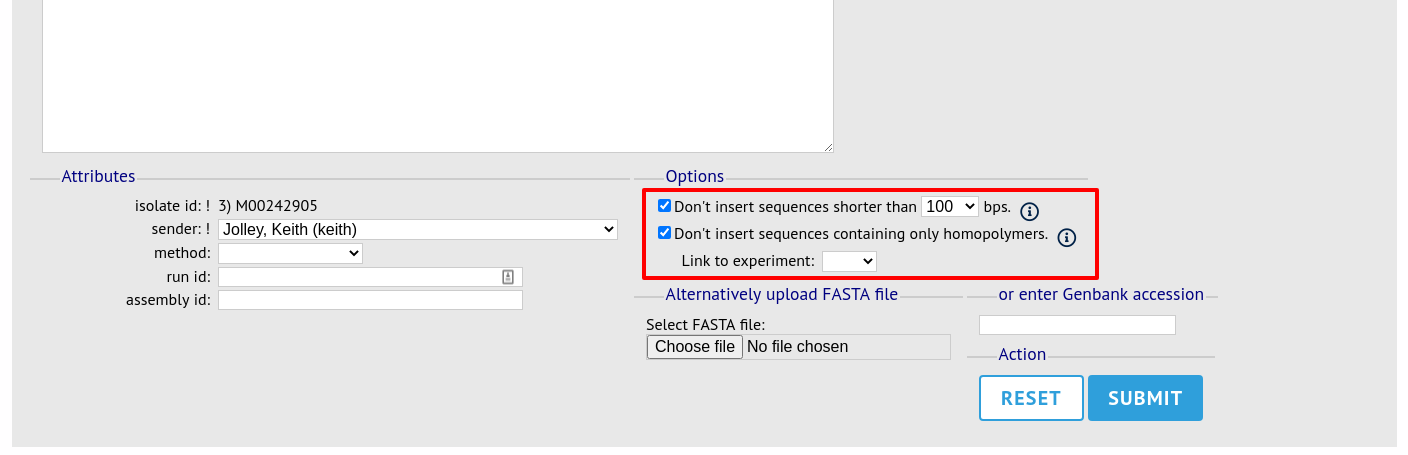
Upload options
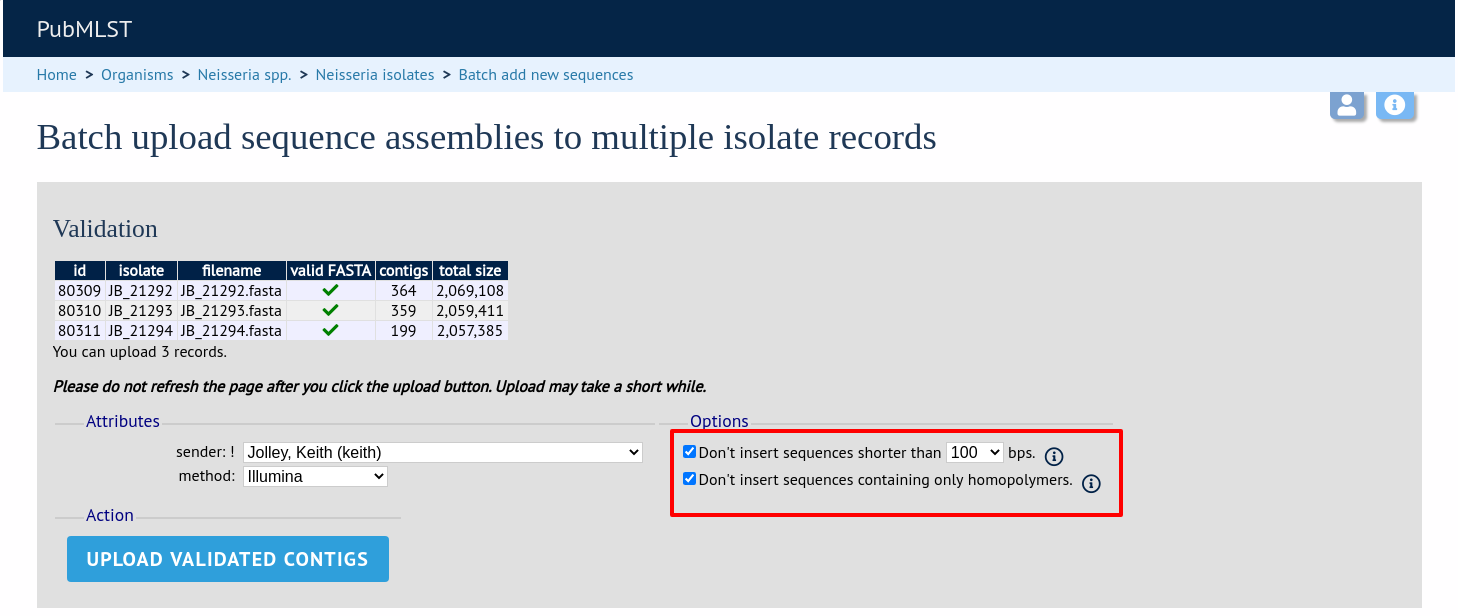
On the upload form, you can select to filter out short sequences or those containing only homopolymeric repeats (which can be artefactually produced by some assembler software versions) from your contig list.
Batch uploading sequence contigs linked to multiple isolate records
To upload contigs for multiple isolates, click the batch add (++) sequence bin link on the curator’s main page.

The first step is to upload the name of the contig file that will be linked to each isolate record. This can be done by pasting two columns in tab-delimited text format (e.g. from a spreadsheet) - the first column contains the isolate identifier, the second contains the filename of the contigs file, which should be in FASTA format.
You can choose which field to use for identifying the isolates, e.g. id (database id) or isolate (name of isolate). The value provided for this field needs to uniquely identify the isolate in the database - please note that only id is guaranteed to be unique.
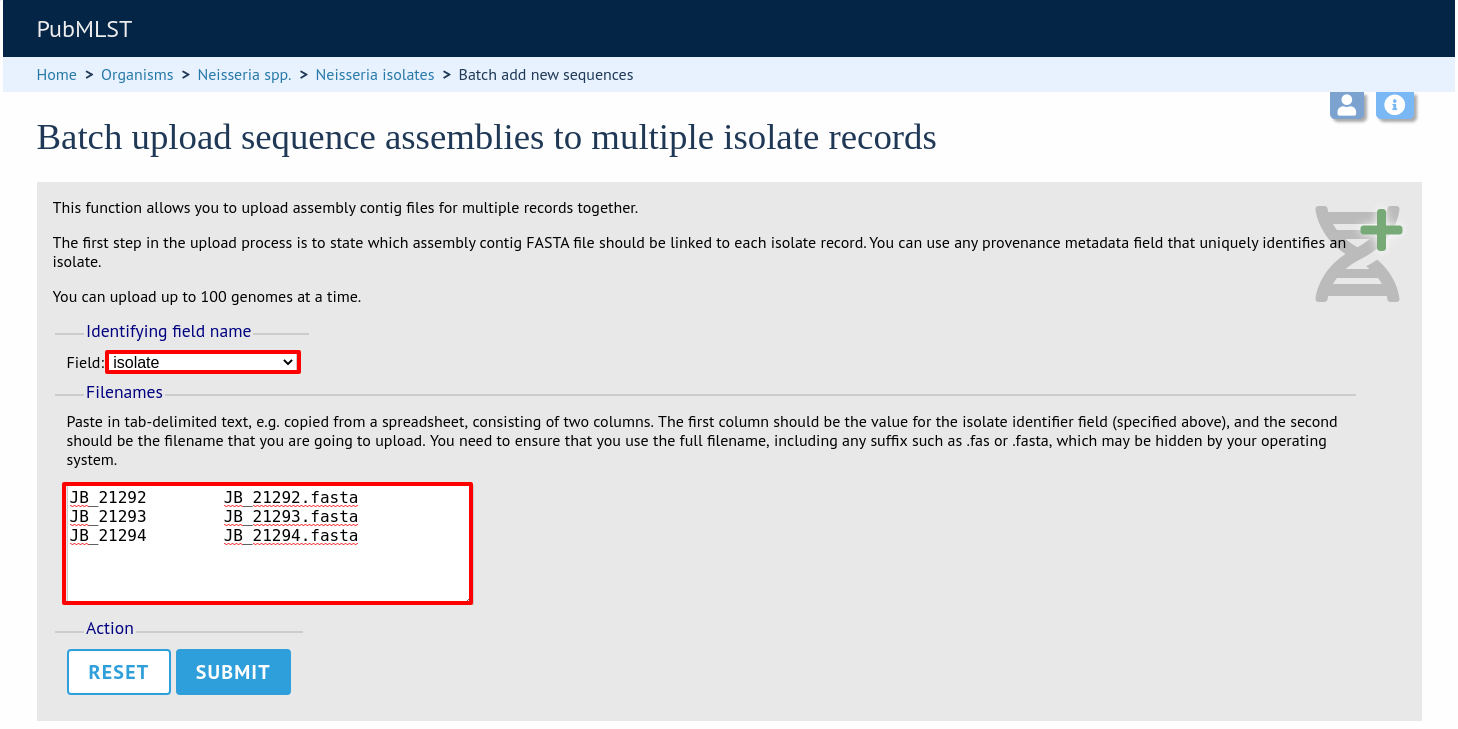
Click Submit. The system will check to make sure that the isolate records are uniquely identified (if not, you will see an error message informing you of this and you will need to use the database id as the identifier). You will then see a file upload form.
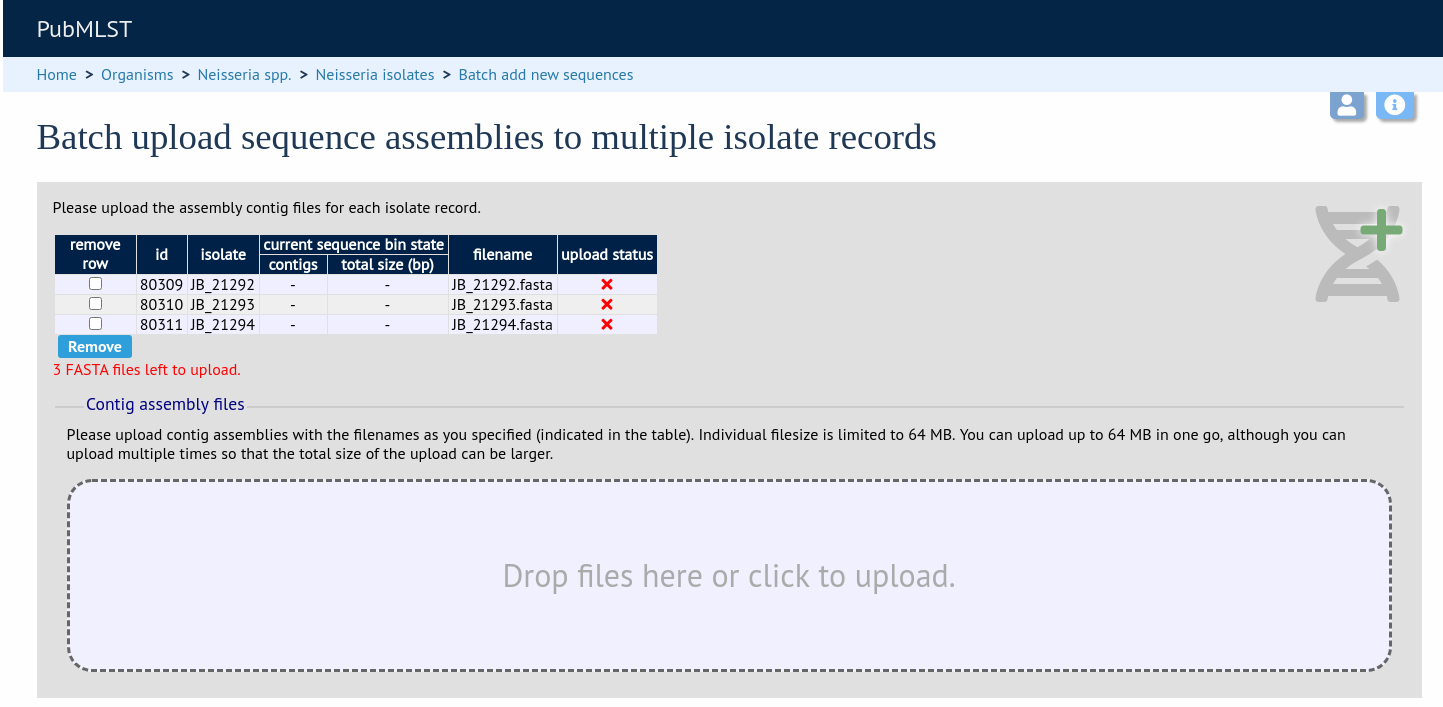
Drag and drop your FASTA format contig files in to the dotted drop area. Provided the filenames exactly match the filename you stated, these will be uploaded to a staging area.
Click ‘Validate’ to check that these files are valid FASTA format.
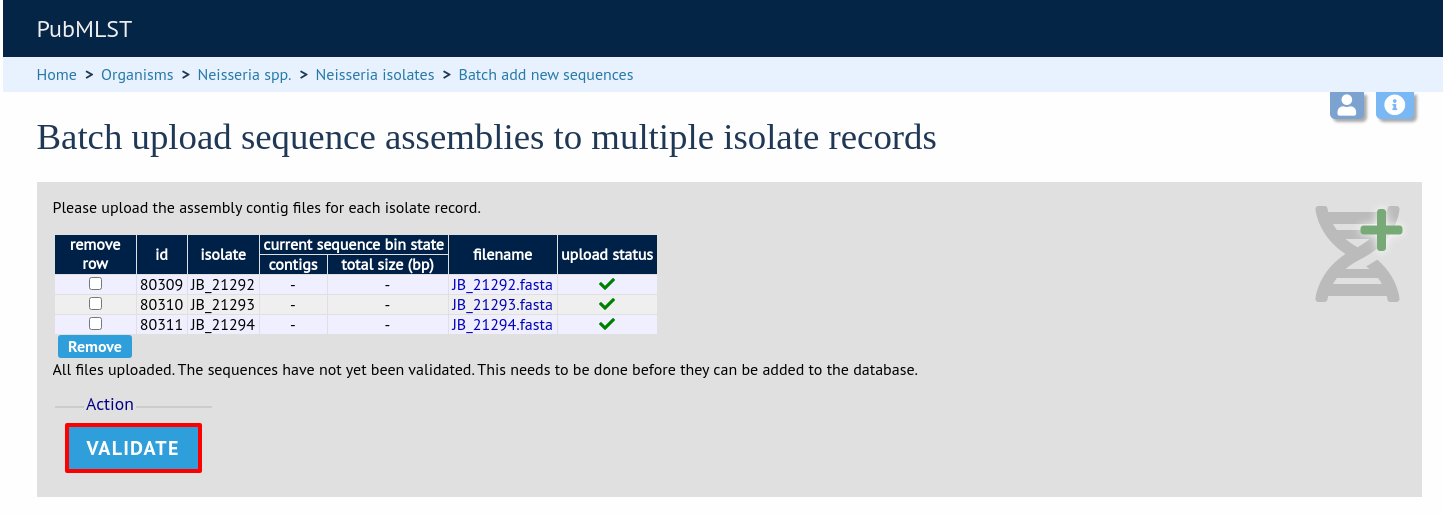
The files will be checked and a table will be displayed showing the total sequence size and number of contigs found. Select the data sender and, optionally the sequencing method from the dropdown lists. Then click ‘Upload validated contigs’.
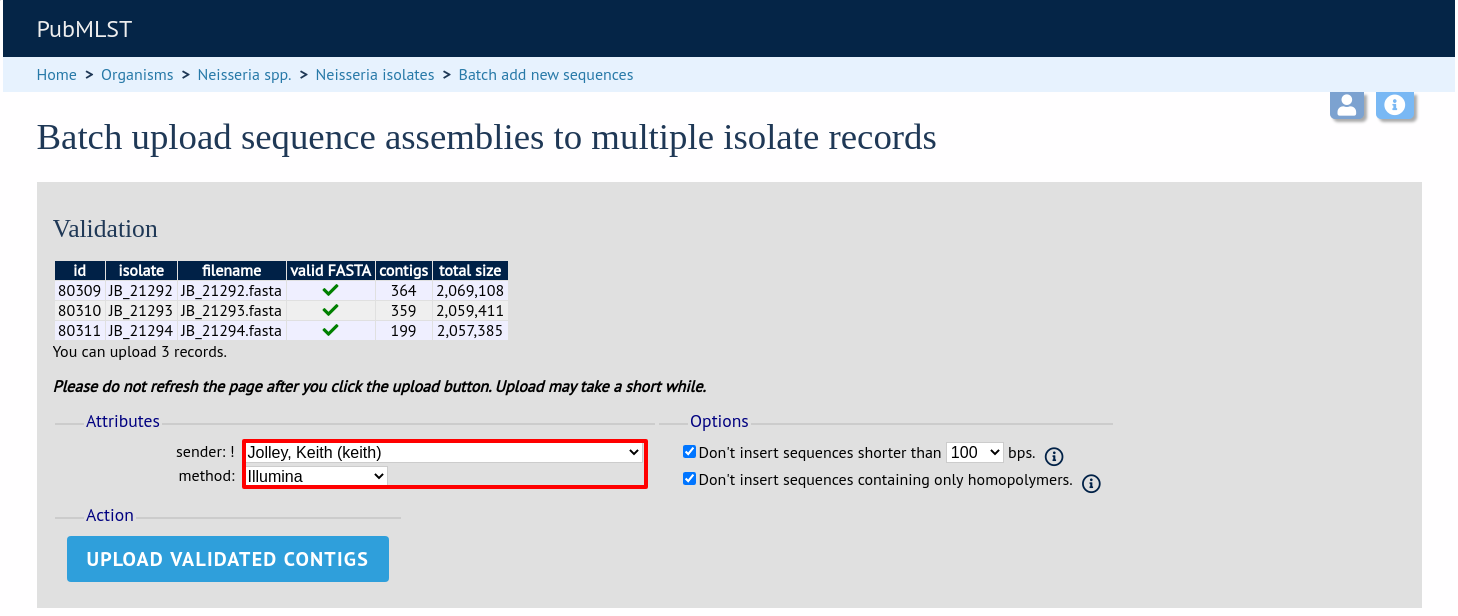
You can also choose to filter out short contigs selecting the checkbox and choosing the minimum length from the dropdown box in the options settings. You can also choose to filter out sequences containing only homopolymeric runs which can be produced artefactually by some assembler versions.
A confirmation message will be displayed after clicking the Upload button.
